Allergy Asthma Immunol Res.
2012 Jan;4(1):31-36. 10.4168/aair.2012.4.1.31.
Involvement of Human Histamine N-Methyltransferase Gene Polymorphisms in Susceptibility to Atopic Dermatitis in Korean Children
- Affiliations
-
- 1Department of Pediatrics and Institute of Allergy, Severance Biomedical Science Institute, BK21 Project for Medical Science, Yonsei University College of Medicine, Seoul, Korea. mhsohn@yuhs.ac
- 2Department of Allergy and Rheumatology, Ajou University School of Medicine, Suwon, Korea.
- KMID: 2130376
- DOI: http://doi.org/10.4168/aair.2012.4.1.31
Abstract
- PURPOSE
Histamine N-methyltransferase (HNMT) catalyzes one of two major histamine metabolic pathways. Histamine is a mediator of pruritus in atopic dermatitis (AD). The aim of this study was to evaluate the association between HNMT polymorphisms and AD in children.
METHODS
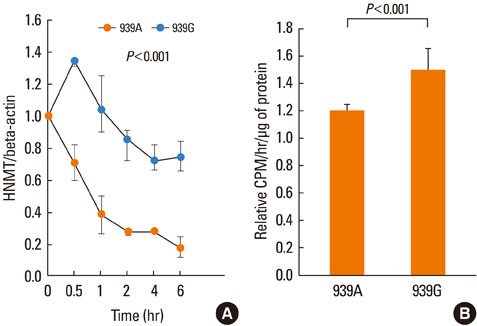
We genotyped 763 Korean children for allelic determinants at four polymorphic sites in the HNMT gene: -465T>C, -413C>T, 314C>T, and 939A>G. Genotyping was performed using a TaqMan fluorogenic 5' nuclease assay. The functional effect of the 939A>G polymorphism was analyzed.
RESULTS
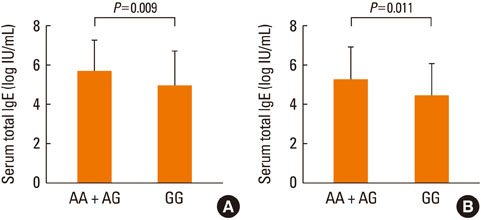
Of the 763 children, 520 had eczema and 542 had atopy. Distributions of the genotype and allele frequencies of the HNMT 314C>T polymorphism were significantly associated with non-atopic eczema (P=0.004), and those of HNMT 939A>G were significantly associated with eczema in the atopy groups (P=0.048). Frequency distributions of HNMT -465T>C and -413C>T were not associated with eczema. Subjects who were AA homozygous or AG heterozygous for 939A>G showed significantly higher immunoglobulin E levels than subjects who were GG homozygous (P=0.009). In U937 cells, the variant genotype reporter construct had significantly higher mRNA stability (P<0.001) and HNMT enzyme activity (P<0.001) than the common genotype.
CONCLUSIONS
Polymorphisms in HNMT appear to confer susceptibility to AD in Korean children.
MeSH Terms
Figure
Cited by 1 articles
-
Overview and challenges of current genetic research on allergic diseases in Korean children
Myunghyun Sohn
Allergy Asthma Respir Dis. 2018;6(Suppl 1):S77-S84. doi: 10.4168/aard.2018.6.S1.S77.
Reference
-
1. Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P, Hamid Q, Kapp A, Leung DY, Lipozencic J, Luger TA, Muraro A, Novak N, Platts-Mills TA, Rosenwasser L, Scheynius A, Simons FE, Spergel J, Turjanmaa K, Wahn U, Weidinger S, Werfel T, Zuberbier T. Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Report. J Allergy Clin Immunol. 2006. 118:152–169.2. Williams H, Flohr C. How epidemiology has challenged 3 prevailing concepts about atopic dermatitis. J Allergy Clin Immunol. 2006. 118:209–213.3. Hong SJ, Ahn KM, Lee SY, Kim KE. The prevalences of asthma and allergic diseases in Korean children. Korean J Pediatr. 2008. 51:343–350.4. Holloway JW, Yang IA, Holgate ST. Genetics of allergic disease. J Allergy Clin Immunol. 2010. 125:S81–S94.5. Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010. 125:16–29.e1-11.6. García-Martín E, Ayuso P, Martínez C, Blanca M, Agúndez JA. Histamine pharmacogenomics. Pharmacogenomics. 2009. 10:867–883.7. Aksoy S, Raftogianis R, Weinshilboum R. Human histamine N-methyltransferase gene: structural characterization and chromosomal location. Biochem Biophys Res Commun. 1996. 219:548–554.8. Wang L, Thomae B, Eckloff B, Wieben E, Weinshilboum R. Human histamine N-methyltransferase pharmacogenetics: gene resequencing, promoter characterization, and functional studies of a common 5'-flanking region single nucleotide polymorphism (SNP). Biochem Pharmacol. 2002. 64:699–710.9. Chen GL, Wang H, Wang W, Xu ZH, Zhou G, He F, Zhou HH. Histamine N-methyltransferase gene polymorphisms in Chinese and their relationship with enzyme activity in erythrocytes. Pharmacogenetics. 2003. 13:389–397.10. García-Martín E, García-Menaya J, Sánchez B, Martínez C, Rosendo R, Agúndez JA. Polymorphisms of histamine-metabolizing enzymes and clinical manifestations of asthma and allergic rhinitis. Clin Exp Allergy. 2007. 37:1175–1182.11. Kim SH, Kang YM, Cho BY, Ye YM, Hur GY, Park HS. Histamine N-methyltransferase 939A>G polymorphism affects mRNA stability in patients with acetylsalicylic acid-intolerant chronic urticaria. Allergy. 2009. 64:213–221.12. Kennedy MJ, Loehle JA, Griffin AR, Doll MA, Kearns GL, Sullivan JE, Hein DW. Association of the histamine N-methyltransferase C314T (Thr105Ile) polymorphism with atopic dermatitis in Caucasian children. Pharmacotherapy. 2008. 28:1495–1501.13. Hanifin JM. Diagnostic criteria for atopic dermatitis: consider the context. Arch Dermatol. 1999. 135:1551.14. Johansson SG, Bieber T, Dahl R, Friedmann PS, Lanier BQ, Lockey RF, Motala C, Ortega Martell JA, Platts-Mills TA, Ring J, Thien F, Van Cauwenberge P, Williams HC. Revised nomenclature for allergy for global use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol. 2004. 113:832–836.15. Preuss CV, Wood TC, Szumlanski CL, Raftogianis RB, Otterness DM, Girard B, Scott MC, Weinshilboum RM. Human histamine N-methyltransferase pharmacogenetics: common genetic polymorphisms that alter activity. Mol Pharmacol. 1998. 53:708–717.16. Yan L, Galinsky RE, Bernstein JA, Liggett SB, Weinshilboum RM. Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000. 10:261–266.17. Deindl P, Peri-Jerkan S, Deichmann K, Niggemann B, Lau S, Sommerfeld C, Sengler C, Müller S, Wahn U, Nickel R, Heinzmann A. No association of histamine- N-methyltransferase polymorphism with asthma or bronchial hyperresponsiveness in two German pediatric populations. Pediatr Allergy Immunol. 2005. 16:40–42.18. Sharma S, Mann D, Singh TP, Ghosh B. Lack of association of histamine-N-methyltransferase (HNMT) polymorphisms with asthma in the Indian population. J Hum Genet. 2005. 50:611–617.19. Darsow U, Pfab F, Valet M, Huss-Marp J, Behrendt H, Ring J, Ständer S. Pruritus and atopic dermatitis. Clin Rev Allergy Immunol. 2011. 41:237–244.20. Petersen J, Drasche A, Raithel M, Schwelberger HG. Analysis of genetic polymorphisms of enzymes involved in histamine metabolism. Inflamm Res. 2003. 52:Suppl 1. S69–S70.21. Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003. 21:579–628.22. Ständer S, Steinhoff M. Pathophysiology of pruritus in atopic dermatitis: an overview. Exp Dermatol. 2002. 11:12–24.23. Kitawaki T, Kadowaki N, Sugimoto N, Kambe N, Hori T, Miyachi Y, Nakahata T, Uchiyama T. IgE-activated mast cells in combination with pro-inflammatory factors induce Th2-promoting dendritic cells. Int Immunol. 2006. 18:1789–1799.24. Reuter S, Stassen M, Taube C. Mast cells in allergic asthma and beyond. Yonsei Med J. 2010. 51:797–807.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Plasma Histamine Levels in Patients with Atopic Dermatitis
- Polymorphisms of the CTLA-4 promoter(-318) and exon 1(+49) genes with atopic dermatitis in Korean children
- Erratum: Involvement of Human Histamine N-Methyltransferase Gene Polymorphisms in Susceptibility to Atopic Dermatitis in Korean Children
- Nipple Involvement in Atopic Dermatitis: Report of 3 cases
- The effect of histamine on the production of interferongamma and interleukin-12 in peripheral blood mononuclear cells from patients with atopic dermatitis