Ann Lab Med.
2014 Nov;34(6):413-425. 10.3343/alm.2014.34.6.413.
Cancer Cytogenetics: Methodology Revisited
- Affiliations
-
- 1Haematology Division, Department of Pathology, The University of Hong Kong, Hong Kong. wantsk@hku.hk
- KMID: 2129566
- DOI: http://doi.org/10.3343/alm.2014.34.6.413
Abstract
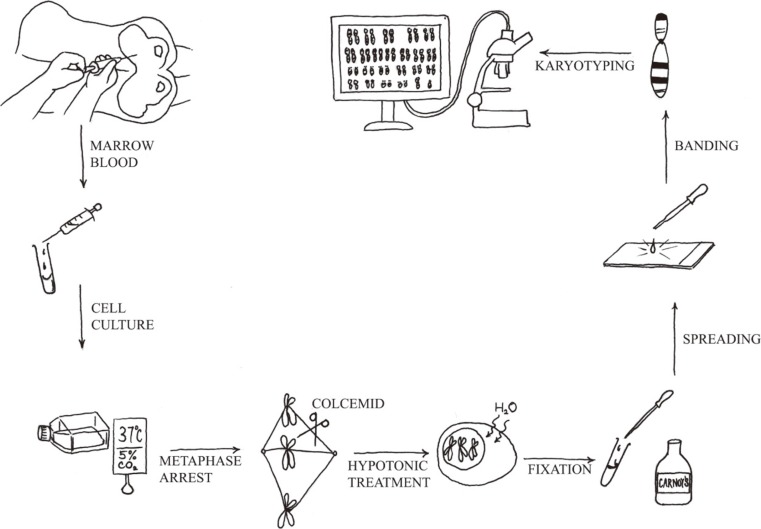
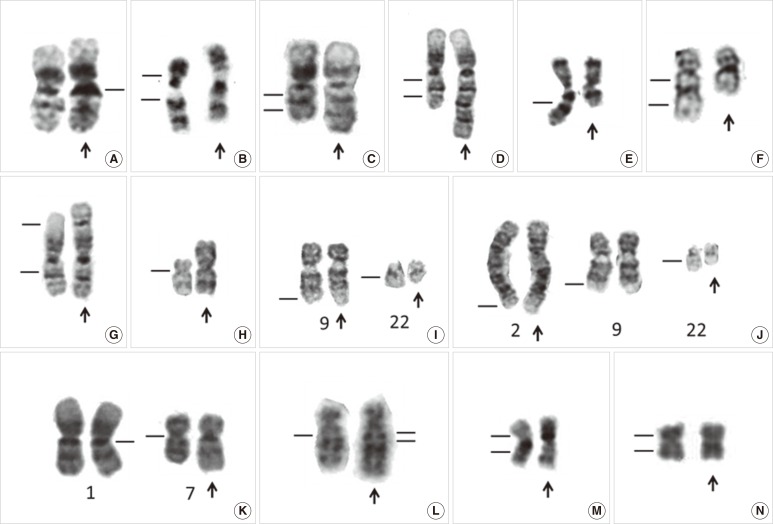
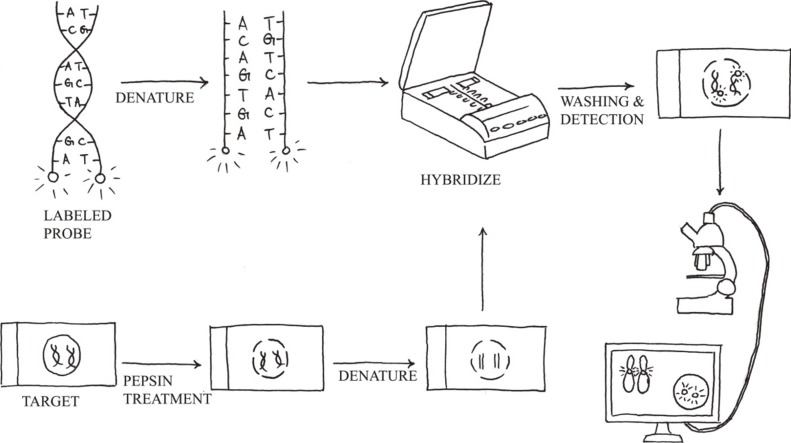
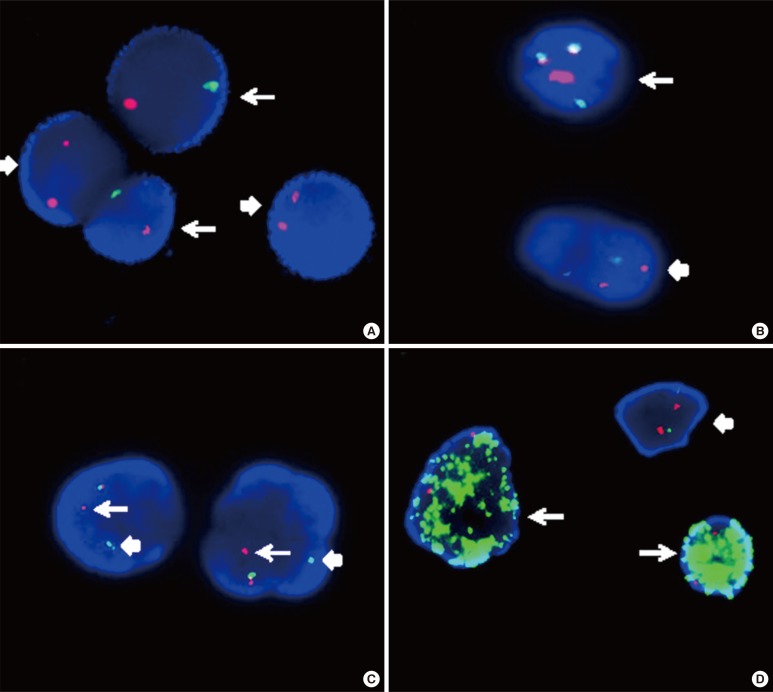
- The Philadelphia chromosome was the first genetic abnormality discovered in cancer (in 1960), and it was found to be consistently associated with CML. The description of the Philadelphia chromosome ushered in a new era in the field of cancer cytogenetics. Accumulating genetic data have been shown to be intimately associated with the diagnosis and prognosis of neoplasms; thus, karyotyping is now considered a mandatory investigation for all newly diagnosed leukemias. The development of FISH in the 1980s overcame many of the drawbacks of assessing the genetic alterations in cancer cells by karyotyping. Karyotyping of cancer cells remains the gold standard since it provides a global analysis of the abnormalities in the entire genome of a single cell. However, subsequent methodological advances in molecular cytogenetics based on the principle of FISH that were initiated in the early 1990s have greatly enhanced the efficiency and accuracy of karyotype analysis by marrying conventional cytogenetics with molecular technologies. In this review, the development, current utilization, and technical pitfalls of both the conventional and molecular cytogenetics approaches used for cancer diagnosis over the past five decades will be discussed.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Anaplastic Large Cell Lymphoma with Massive Eosinophilia and Complex Karyotype Initially Misdiagnosed as Chronic Eosinophilic Leukemia
Min-Kyung So, Sholhui Park, Min-Sun Cho, Yeung Chul Mun, Jungwon Huh
Lab Med Online. 2018;8(2):56-61. doi: 10.3343/lmo.2018.8.2.56.
Reference
-
1. Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960; 132:1497.2. Rowley JD. A new consistent chromosomal abnormality in chronic myelogeneous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973; 243:290–293. PMID: 4126434.3. de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, et al. A cellular oncogene is translocated to Philadelphia chromosome in chronic myelocytic leukemia. Nature. 1982; 300:765–767. PMID: 6960256.4. Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasm and acute leukemia: rationale and important changes. Blood. 2009; 114:937–951. PMID: 19357394.5. Manuelidis L, Langer-Safer PR, Ward DC. High-resolution mapping of satellite DNA using biotin-labeled DNA probes. J Cell Biol. 1982; 95:619–625. PMID: 6754749.
Article6. Wan TS, Ma ES. Molecular cytogenetics: an indispensable tool for cancer diagnosis. Chang Gung Med J. 2012; 35:96–110. PMID: 22537925.7. Wan TS, Ma ES. The role of FISH in hematologic cancer. Int J Hematol Oncol. 2012; 1:71–86.
Article8. Wan TS, Ma SK, Chan GC, Ching LM, Ha SY, Chan LC. Complex cytogenetic abnormalities in T-lymphoblastic lymphoma: resolution by spectral karyotyping. Cancer Genet Cytogenet. 2000; 118:24–27. PMID: 10731586.9. Raza A, Maheshwari Y, Preisler HD. Differences in cell cycle characteristics among patients with acute nonlymphocytic leukemia. Blood. 1987; 69:1647–1653. PMID: 3555651.
Article10. Yunis JJ. Comparative analysis of high-resolution chromosome techniques for leukemic bone marrows. Cancer Genet Cytogenet. 1982; 7:43–50. PMID: 6754071.
Article11. Garipidou V, Secker-Walker LM. The use of fluorodeoxyuridine synchronization for cytogenetic investigation of acute lymphoblastic leukemia. Cancer Genet Cytogenet. 1991; 52:107–111. PMID: 1826229.
Article12. Brothman AR, Schneider NR, Saikevych I, Cooley LD, Butler MG, Patil S, et al. Cytogenetic Heteromorphisms. Survey results and reporting practices of Giemsa-band regions that we have pondered for years. Arch Pathol Lab Med. 2006; 130:947–949. PMID: 16831047.
Article13. Wan TS, Ma SK, Chan LC. Acquired pericentric inversion of chromosome 9 in essential thrombocythemia. Hum Genet. 2000; 106:669–670. PMID: 10942117.
Article14. Lee SG, Park TS, Lim G, Lee KA, Song J, Choi JR. Constitutional pericentric inversion 9 and hematological disorders: a Korean tertiary institution's experience over eight years. Ann Clin Lab Sci. 2010; 40:273–277. PMID: 20689141.15. Wan TS, Au WY, Chan JC, Chan LC, Ma SK. Trisomy 21 as the sole acquired karyotypic abnormality in acute myeloid leukemia and myelodysplastic syndrome. Leuk Res. 1999; 23:1079–1083. PMID: 10576514.
Article16. Wan TS, Ma SK, Au WY, Liu HS, Chan JC, Chan LC. Trisomy 21 and other chromosomal abnormalities in acute promyelocytic leukemia. Cancer Genet Cytogenet. 2003; 140:170–173. PMID: 12645658.
Article17. Ma SK, Wan TS, Cheuk AT, Fung LF, Chan GC, Chan SY, et al. Characterization of additional genetic events in childhood acute lymphoblastic leukemia with TEL/AML1 gene fusion: a molecular cytogenetics study. Leukemia. 2001; 15:1442–1447. PMID: 11516105.
Article18. Ma SK, Wan TS. Single autosomal trisomy in acute myeloid leukemia and myelodysplastic syndrome. Curr Genomics. 2000; 1:153–173.
Article19. Heim S, Mitelman F. Numerical chromosome aberrations in human neoplasia. Cancer Genet Cytogenet. 1986; 22:99–108. PMID: 3708552.
Article20. Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004; 432:338–341. PMID: 15549096.
Article21. Ma SK, Wan TS, Au EY, Chan LC. Trisomy 5 in two cases of acute monocytic leukemia with hyperdiploid clones. Leuk Res. 1998; 22:961–964. PMID: 9766757.
Article22. Wan TS, Yip SF, Yeung YM, Chan LC, Ma SK. Fatal diffuse alveolar damage complicating acute myeloid leukemia with abnormal eosinophils and trisomy X. Ann Hematol. 2002; 81:167–169. PMID: 11904745.
Article23. Mehta AB, Bain BJ, Fitchett M, Shah S, Secker-Walker LM. Trisomy 13 and myeloid malignancy-characteristic blast cell morphology: a United Kingdom Cancer Cytogenetics Group survey. Br J Haematol. 1998; 101:749–752. PMID: 9674750.24. Ma SK, Wan TS. Blast cell morphology in acute myeloid leukemia with trisomy 13. Leuk Res. 1999; 23:767–769. PMID: 10456675.25. Yip SF, Wan TS, Chan LC, Chan GC. Trisomy 4 as sole karyotypic abnormality in acute lymphoblastic leukemia: different clinical features and treatment response between B and T phenotypes? Cancer Genet Cytogenet. 2006; 164:94–95. PMID: 16364773.
Article26. Au WY, Ma SK, Wan TS, Jim MH, Kwong YL. Subvalvular pulmonary stenosis, demyelination and myelodysplasia with monosomy 7. Leuk Lymphoma. 2002; 43:1505–1507. PMID: 12389638.
Article27. Au WY, Wan TS, Leung RY, Lie AK. Sequential chronic myelogenous leukemia, B-lineage lymphoma and erythroleukemia with monosomy 7 over 10 years. Leuk Lymphoma. 2012; 53:733–735. PMID: 21958131.
Article28. Surapolchai P, Ha SY, Chan GC, Lukito J, Wan TS, So CC, et al. Central diabetes insipidus: an unusual complication in a child with juvenile myelomonocytic leukemia and monosomy 7. J Pediatr Hematol Oncol. 2013; 35:e84–e87. PMID: 22858568.29. Wan TS, Ma ES, Lam CC, Chan LC, Lee KK, Au WY. Deletion 9q as the sole karyotypic abnormality in myelocytic disorders: a new case of myelodysplastic syndrome and its prognostic implications in acute myelocytic leukemia. Cancer Genet Cytogenet. 2003; 145:184–186. PMID: 12935935.
Article30. Wan TS, Chim CS, So CK, Chan LC, Ma SK. Complex variant 15;17 translocations in acute promyelocytic leukemia. A case report and review of three-way translocations. Cancer Genet Cytogenet. 1999; 111:139–143. PMID: 10347551.31. So CW, Ma SK, Wan TS, Chan GC, Ha SY, Chan LC. Analysis of MLL-derived transcripts in infant acute monocytic leukemia with a complex translocation (1;11;4)(q21;q23;p16). Cancer Genet Cytogenet. 2000; 117:24–27. PMID: 10700861.
Article32. So CC, Wan TS, Yip SF, Chan LC. A dual colour dual fusion fluorescence in situ hybridization study on the genesis of complex variant translocations in chronic myelogenous leukemia. Oncol Rep. 2008; 19:1181–1184. PMID: 18425374.33. Wan TS, So CC, Hui KC, Yip SF, Ma ES, Chan LC. Diagnostic utility of dual fusion PML/RARα translocation DNA probe (D-FISH) in acute promyelocytic leukemia. Oncol Rep. 2007; 17:799–805. PMID: 17342318.
Article34. Wan TS, Ma SK, Yip SF, Yeung YM, Chan LC. Molecular characterization of der(15)t(11;15) as a secondary cytogenetic abnormality in acute promyelocytic leukemia with cryptic PML-RARα fusion on chromosome 17q. Cancer Genet Cytogenet. 2000; 121:90–93. PMID: 10958948.35. So CC, Wan TS, Ma ES, Chan LC. An unbalanced translocation, der(17)t(1;17)(p13;p11.2), leads to heterozygous loss of TP53 and is associated with clinical evolution in myelodysplastic syndrome. Br J Biomed Sci. 2008; 65:36–38. PMID: 18476495.
Article36. Willem P, Pinto M, Bernstein R. Translocation t(1;7) revisited. Report of three further cases and review. Cancer Genet Cytogenet. 1988; 36:45–54. PMID: 3060251.37. So CC, Ma ES, Wan TS, Yip SF, Chan LC. Clinicopathological features of unbalanced translocation Der(1;7)(q10;p10) in myeloid neoplasms. Leuk Res. 2008; 32:1000–1001. PMID: 17980911.
Article38. Pedersen B, Nørgaard JM, Pedersen BB, Clausen N, Rasmussen IH, Thorling K. Many unbalanced translocation show duplication of a translocation participant. Clinical and cytogenetic implications in myeloid hematologic malignancies. Am J Hematol. 2000; 64:161–169. PMID: 10861810.39. Sanada M, Uike N, Ohyashiki K, Ozawa K, Lili W, Hangaishi A, et al. Unbalanced translocation der(1;7)(q10;p10) defines a unique clinicopathological subgroup of myeloid neoplasms. Leukemia. 2007; 21:992–997. PMID: 17315020.
Article40. Wan TS, Ma SK, Li CK, Chan LC. Atypical fluorescence in situ hybridization pattern in chronic myeloid leukemia due to cryptic insertion of BCR at 9q34. Leukemia. 2004; 18:161–162. PMID: 14603334.41. So CC, Wan TS, Chow JL, Hui KC, Choi WW, Lam CC, et al. A single-center cytogenetic study of 629 Chinese patients with de novo acute myeloid leukemia – evidence of major ethnic differences and a high prevalence of acute promyelocytic leukemia in Chinese patients. Cancer Genet. 2011; 204:430–438. PMID: 21962893.42. Cheng Y, Wang Y, Wang H, Chen Z, Lou J, Xu H, et al. Cytogenetic profile of de novo acute myeloid leukemia: a study based on 1432 patients in a single institution of China. Leukemia. 2009; 23:1801–1806. PMID: 19474801.
Article43. Saxe DF, Persons DL, Wolff DJ, Theil KS. Cytogenetics Resource Committee of the College of American Pathologists. Validation of fluorescence in situ hybridization using an analyste-specific reagent for detection of abnormalities involving the mixed lineage leukemia gene. Arch Pathol Lab Med. 2012; 136:47–52. PMID: 22208487.44. American College of Medical Genetics. Standards and Guidelines for Clinical Genetics Laboratories. https://www.acmg.net/StaticContent/SGs/Section_E_2011.pdf (2009 edition, Revised on Jan 2010).45. Shaffer LG, McGowan-Jordan J, Schmid M, editors. ISCN (2013): An International System for Human Cytogenetic Nomenclature. Basel: S. Karger;2013.46. Ross FM, Avet-Loiseau H, Ameye G, Gutiérrez NC, Liebisch P, O'Connor S, et al. Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica. 2012; 97:1272–1277. PMID: 22371180.
Article47. Ma SK, Wan TS, Au WY, Kwong YL, Chan LC. Atypical chronic myeloid leukemia with der(20)t(17;20)(q21;q13). Cancer Genet Cytogenet. 1999; 112:130–133. PMID: 10686939.
Article48. Wan TS, Ma SK, Yip SF, Yeung YM, Chan LC. Two balanced and novel chromosomal translocations in myeloid malignancies.characterization by multiplex fluorescence in situ hybridization. Cancer Genet Cytogenet. 2002; 139:52–56. PMID: 12547159.49. Ma SK, Lee AC, Wan TS, Lam CK, Chan LC. Trisomy 8 as a secondary genetic change in acute megakaryoblastic leukemia associated with Down's syndrome. Leukemia. 1999; 13:491–492. PMID: 10086746.
Article50. Ma SK, Kwong YL, Shek TW, Wan TS, Chow EY, Chan JC, et al. The role of trisomy 8 in the pathogenesis of chronic eosinophilic leukemia. Hum Pathol. 1999; 30:864–868. PMID: 10414507.
Article51. Cheung AM, Wan TS, Leung JC, Chan LY, Huang H, Kwong YL, et al. Aldehyde dehydrogenase activity in leukemia blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007; 21:1423–1430. PMID: 17476279.52. Meyer C, Kowarz E, Yip SF, Wan TS, Chan TK, Dingermann T, et al. A complex MLL rearrangement identified five years after initial MDS diagnosis results in out-of-frame fusions without progression to acute leukemia. Cancer Genet. 2011; 204:557–562. PMID: 22137486.
Article53. Ma SK, Wan TS, Au WY, Fung LF, So CK, Chan LC. Chromosome 11q deletion in myeloid malignacies. Leukemia. 2002; 16:953–955. PMID: 11986961.54. Cheung AM, Fung TK, Fan AK, Wan TS, Chow HC, Leung JC, et al. Successful engraftment by leukemia initiating cells in adult acute lymphoblastic leukemia after direct intrahepatic injection into unconditioned newborn NOD/SCID mice. Exp Hematol. 2010; 38:3–10. PMID: 19837128.
Article55. Wan TS, Ma ES, Chen YT. Near-tetraploid acute myeloid leukemia. Br J Haematol. 2011; 155:285. PMID: 21679166.56. So CC, Yung KH, Chu ML, Wan TS. Diagnostic challenges in a case of B cell lymphoma unclassifiable with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma. Int J Hematol. 2013; 98:478–482. PMID: 23959583.
Article57. Lee JH, Wan TS, Ha JS. Acute myeloid leukemia with a novel t(8;21) variant: paracentric inversion-associatedins(21;8). Leuk Lymphoma. 2014; 55:441–443. PMID: 23672346.58. Li YH, Ma SK, Wan TS, Au WY, Fung LF, Leung AY. Lineage-specific differences in telomere length after bone marrow transplantation. Bone Marrow Transplant. 2002; 30:475–477. PMID: 12368963.
Article59. Yang JJ, Marschalek R, Meyer C, Park TS. Diagnostic usefulness of genomic breakpoint analysis of various gene rearrangements in acute leukemias: a perspective of long distance- or long distance inverse-PCR-based approaches. Ann Lab Med. 2012; 32:316–318. PMID: 22779077.
Article60. Wan TS, Ma ES, Chan GC, Chan LC. Investigation of MYCN status in neuroblastoma by fluorescence insitu hybridization. Int J Mol Med. 2004; 14:981–987. PMID: 15547663.61. Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Compararive genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992; 258:818–821. PMID: 1359641.62. Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996; 273:494–497. PMID: 8662537.
Article63. Speicher MR, Gwyn Ballard S, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet. 1996; 12:368–375. PMID: 8630489.
Article64. Chudoba I, Plesch A, Lörch T, Lemke J, Claussen U, Senger G. High resolution multicolor-banding: a new technique for refine FISH analysis of human chromosomes. Cytogenet Cell Genet. 1999; 84:156–160. PMID: 10393418.65. Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genetics. 1998; 20:207–211. PMID: 9771718.
Article66. Tsao SW, Wong N, Wang X, Liu Y, Wan TS, Fung LF, et al. Nonrandom chromosomal imbalances in human ovarian surface epithelial cells immortalized by HPV16-E6E7 viral oncogenes. Cancer Genet Cytogenet. 2001; 130:141–149. PMID: 11675135.
Article67. Hu YC, Lam KY, Law SY, Wan TS, Ma ES, Kwong YL, et al. Establishment, characterization, karyotyping, and comparative genomic hybridization analysis of HKESC-2 and HKESC-3, two newly established human esophageal squamous cell carcinoma cell lines. Cancer Genet Cytogenet. 2002; 135:120–127. PMID: 12127396.68. Wong MP, Fung LF, Wang E, Chow WS, Chiu SW, Lam WK, et al. Chromosomal aberrations of primary lung adenocarcinomas in nonsmokers. Cancer. 2003; 97:1263–1270. PMID: 12599234.
Article69. Peeper D, Berns A. Cross-species oncogenomics in cancer gene identification. Cell. 2006; 125:1230–1233. PMID: 16814709.
Article70. Slovak ML, Bedell V, Hsu YH, Estrine DB, Nowak NJ, Delioukina ML, et al. Molecular karyotypes of Hodgkin and Reed/Sternberg cells at disease onset reveal distinct copy number alterations in chemosensitive versus refractory Hodgkin lymphoma. Clin Cancer Res. 2011; 17:3443–3454. PMID: 21385932.
Article71. Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009; 106:12950–12955. PMID: 19651600.
Article72. Yu L, Slovak ML, Mannoor K, Chen C, Hunger SP, Carroll AJ, et al. Microarray detection of multiple recurring submicroscopic chromosomal aberrations in pediatric T-cell acute lymphoblastic leukemia. Leukemia. 2011; 25:1042–1046. PMID: 21383747.
Article73. Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008; 111:776–784. PMID: 17890455.
Article74. O'Keefe C, McDevitt MA, Maciejewski JP. Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies. Blood. 2010; 115:2731–2739. PMID: 20107230.75. Cooley LD, Lebo M, Li MM, Slovak ML, Wolff DJ. Working Group of the American College of Medical Genetics and Genomics (ACMG) Laboratory Quality Assurance Committee. American College of Medical Genetics and Genomics technical standards and guidelines: microarray analysis for chromosome abnormalities in neoplastic disorders. Genet Med. 2013; 15:484–494. PMID: 23619274.
Article76. Mitelman F, Johansson B, Mertens F, editors. Mitelman database of chromosome aberrations in cancer. Updated on May 2014. http://cgap.nci.nih.gov/Chromosomes/Mitelman.77. Atlas of Genetics and Cytogenetics in Oncology and Haematology. Updated on May 2012. http://AtlasGeneticsOncology.org.78. Huret JL, Ahmad M, Arsaban M, Bernheim A, Cigna J, Desangles F, et al. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013; 41(Database issue):D920–D924. PMID: 23161685.
Article79. Shaffer LG, Tommerup N, editors. ISCN (2005): an International System for Human Cytogenetic Nomenclature. Basel: S. Karger;2005.80. Mitelman F, Rowley JD. ISCN (2005) is not acceptable for describing clonal evolution in cancer. Genes Chromosomes Cancer. 2007; 46:213–214. PMID: 17171683.
Article81. Mascarello JT, Cooley LD, Davison K, Dewald GW, Brothman AR, Herrman M, et al. Problems with ISCN FISH Nomenclature make it not practical for use in clinical test reports or cytogenetic databases [corrected]. Genet Med. 2003; 5:370–377. PMID: 14501832.82. Shaffer LG, Slovak ML, Campbell LJ, editors. ISCN (2009): an International System for Human Cytogenetic Nomenclature. Basel: S. Karger;2009.83. Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, et al. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996; 5:685–691. PMID: 8733138.
Article84. Wan TS, Martens UM, Poon SS, Tsao SW, Chan LC, Lansdorp PM. Absence or low number of telomere repeats at junctions of dicentric chromosomes. Genes Chromosomes Cancer. 1999; 24:83–86. PMID: 9892113.
Article85. Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, et al. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci USA. 1997; 94:7423–7428. PMID: 9207107.
Article86. Yamada NA, Rector LS, Tsang P, Carr E, Scheffer A, Sederberg MC, et al. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet Genome Res. 2011; 132:248–254. PMID: 21178330.
Article87. Stevens-Kroef MJ, van den Berg E, Olde Weghuis D, Geurts van Kessel A, Pfundt R, Linssen-Wiersma M, et al. Identification of prognostic relevant chromosomal abnormalities in chronic lymphocytic leukemia using microarray-based genomic profiling. Mol Cytogenet. 2014; 7:3. PMID: 24401281.
Article88. Hallek M, Bergsagel PL, Anderson KC. Multiple myeloma: increasing evidence for a multistep transformation process. Blood. 1998; 91:3–21. PMID: 9414264.
Article89. Leung EW, Sin PL, Wan TS. The impact of FICTION on the detection of genetic aberrations in multiple myeloma. J H K Inst Med Lab Sci. 2012; 13:1–8.