J Bacteriol Virol.
2006 Jun;36(2):79-87. 10.4167/jbv.2006.36.2.79.
Genetic Variation in the NSP4 Gene of Human Rotavirus Isolated in Seoul
- Affiliations
-
- 1Department of Microbiology, Chung-Ang University College of Medicine, Korea. kimwy@cau.ac.kr
- 2Department of Pediatrics, Chung-Ang University College of Medicine, Korea.
- KMID: 2055014
- DOI: http://doi.org/10.4167/jbv.2006.36.2.79
Abstract
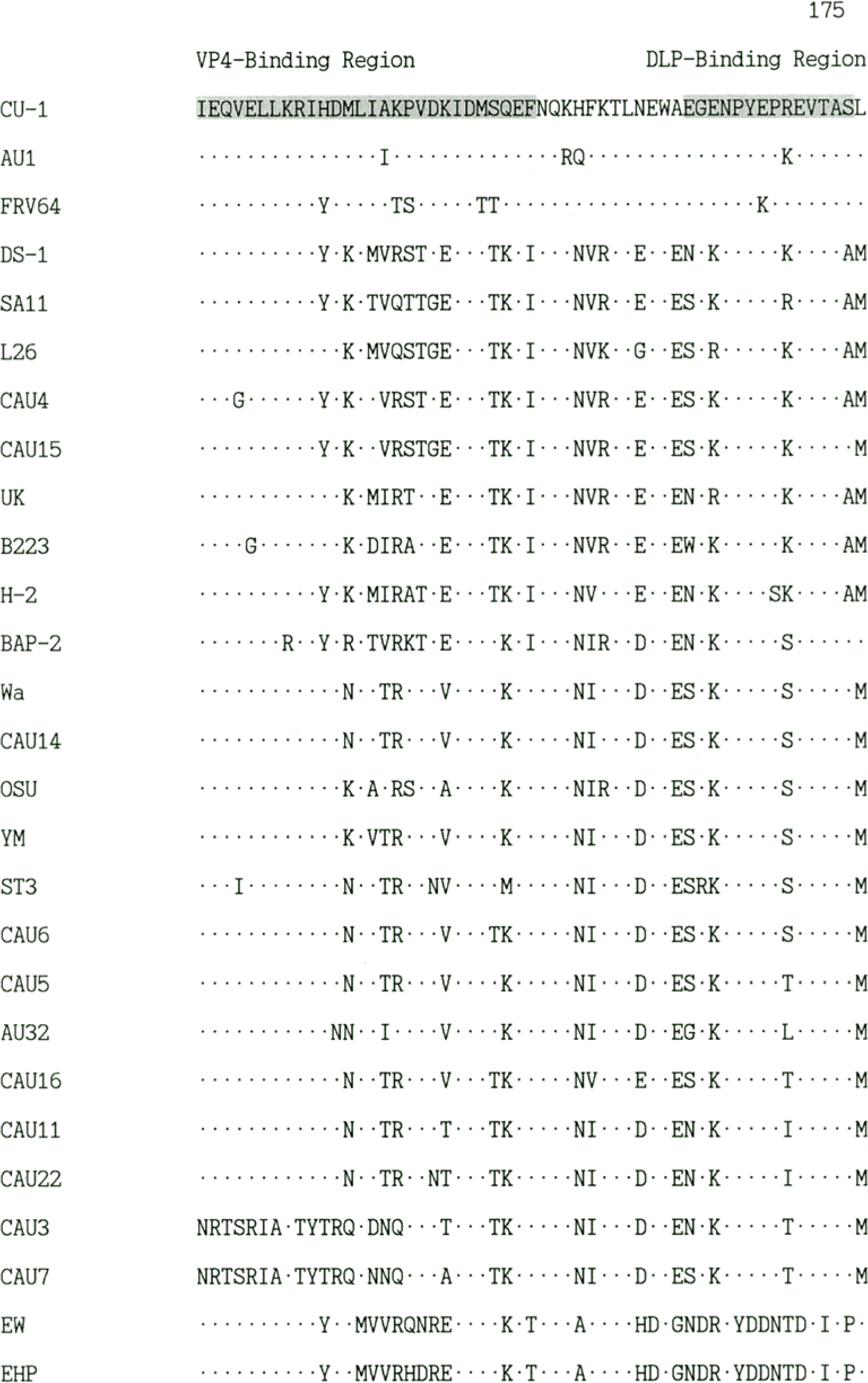
- The nonstructural protein 4 (NSP4) of rotavirus encoded by gene 10, plays an important role in rotavirus pathogenicity. In this study, NSP4 gene sequences of human rotaviruses circulating in Seoul, Korea between March 2004 and April 2005 were determined. The nucleotide sequence data indicated that the NSP4 genes of human rotavirus Korean isolates were 750 or 751 bases in length and encoded one open reading frame of 175 amino acids with two glycosylation sites. The NSP4 of Korean isolates exhibited amino acid sequence homologies between 59.4% and 98.9%. The NSP4 of CAU4 and CAU15 showed a high degree of amino acid sequence homologies with NSP4 genotype A viruses, but the NSP4 of CAU5, CAU6, CAU11, CAU14, CAU16 and CAU22 exhibited a high degree of amino acid sequence homologies with NSP4 genotype B viruses. Interestingly, CAU3 and CAU7 showed low degree of amino acid sequence homology with those of currently described NSP4 genotypes A to D and belonged a distinct lineage on the phylogenetic tree. These findings suggests that distinct NSP4 type was circulating among human rotavirus strains in the local community of Seoul and raising intriguing questions regarding possible explanations for new genotype.
Keyword
MeSH Terms
Figure
Reference
-
References
1). Au KS, Mattion NM, Estes MK. A subviral particle binding domain on the rotavirus nonstructural glycoprotein NS28. Virology. 194:665–673. 1993.
Article2). Ball JM, Tian P, Zeng CQY, Morris AP, Estes MK. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 272:101–104. 1996.
Article3). Ballard A, McCrae MA, Desselberger U. Nucleotide sequences of normal and rearranged RNA segments 10 of human rotaviruses. J Gen Virol. 73:633–638. 1992.
Article4). Bergmann CC, Maass D, Pourchynsky MS. Topology of the non-structural rotavirus receptor glycoprotein in NS28 in the rough endoplasmic reticulum. EMBO J. 8:1695–1703. 1989.5). Bresee J, Fang ZY, Wang B, Nelson EA, Tam J, Soenarto Y, Wilopo SA, Kilgore P, Kim JS, Kang JO, Lan WS, Gaik CL, Moe K, Chen KT, Jiraphongsa C, Ponguswanna Y, Nguyen VM, Phan VT, Le TL, Hummelman E, Gentsch JR, Glass R. First report from the Asian Rotavirus Surveillance Network. Emerg Infect Dis. 10:988–995. 2004.
Article6). CDMR. Viral Agents of Gastroenteritis in 2003. http://dis.mohw.go.kr/cdmr/cdmr_view.asp. 2004.7). Ciarlet M, Liprandi F, Conner ME, Estes MK. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analysis of animal rota-viruses. Arch Virol. 145:371–383. 2000.8). Estes MK. Advances in molecular biology: impact on rota-virus vaccine development. J Infect Dis. 174(Suppl. 1):S37–S46. 1996.
Article9). Estes MK, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 53:410–449. 1989.
Article10). Felsenstein J. PHYLIP (Phylogeny Inference Package) vession 3.5c. Department of Genetics, University of Washington, Seattle. 1993.11). Gouvea V, de Castro L, Timenetsky MC, Greenberg H, Santos N. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J Clin Microbiol. 32:1408–1409. 1994.
Article12). Jukes TH, Cantor CR. Evolution of protein molecules. pp.p. 21–132. In. Mammalian Protein Metabolism. volume. 3:Munro HN, editor. (Ed),. Academic Press Inc;New York: 1969.
Article13). Kirwood CD, Palombo EA. Genetic characterization of the rotavirus nonstructural protein, NSP4. Virology. 236:258–265. 1997.
Article14). Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mole Biol Evol. 4:406–425. 1987.15). Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows Interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acid Res. 24:4876–4882. 1997.16). Tian P, Ball JM, Zeng CQY, Estes MK. The rotavirus non-structural glycoprotein NSP4 possesses membrane destabilization activity. J Virol. 70:6973–6981. 1996.
Article17). World Health Organization. Rotavirus vaccine. WHO WER. 74:33–38. 1999.18). Zang M, Zeng CQY, Dong Y, Ball JM, Saif LJ, Morris AP, Estes MK. Mutations in rotavirus nonstructural glycoprotein NSP4 are associated with altered virus virulence. J Virol. 72:3666–3672. 1998.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Cloning and Sequencing of Nonstructural Protein (NSP4) Gene of Bovine Rotavirus Isolated in Korea
- Analysis of structure-function relationship in porcine rotavirus A enterotoxin gene
- Expression of Nonstructural Protein 4 (NSP4) of Bovine Rotavirus Isolated in Korea
- Isolation and Characterization of G9 Human Rotaviruses
- Role of Rotavirus Enterotoxin NSP4 in the Inflammatory Response in Murine Macrophage RAW 264.7 Cells