Cancer Res Treat.
2004 Feb;36(1):6-12.
p63 and p73 in Tumor Suppression and Promotion
- Affiliations
-
- 1Department of Cell Biology, Harvard Medical School, USA.
Abstract
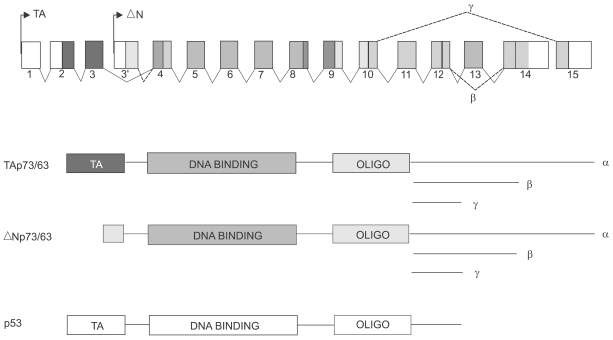
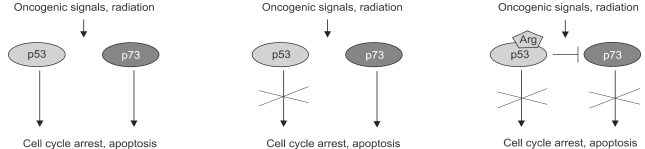
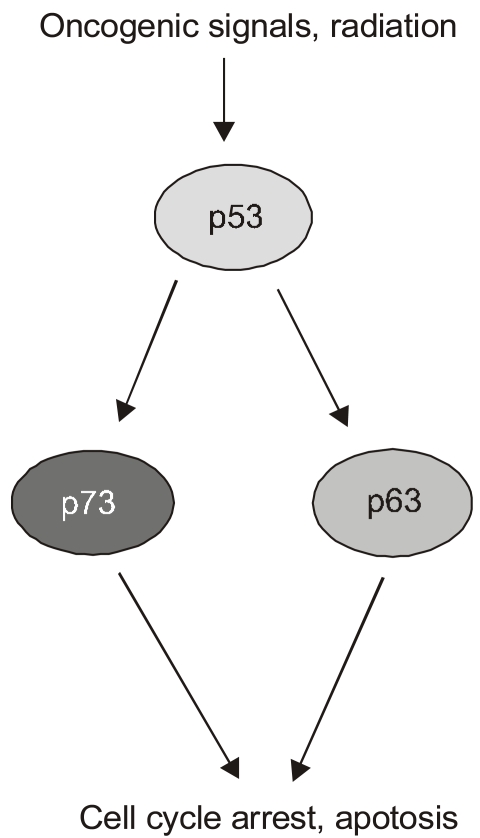
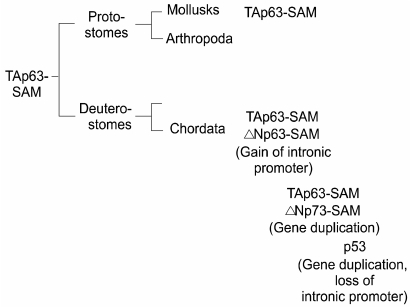
- The recent discovery of two genes, termed p63 and p73, encoding transcription factors highly homologous to p53 presents unexpected challenges and opportunities for the understanding and treatment of cancers. The questions raised are many but center on determining whether these new genes possess novel tumor suppressor functions, cooperate with p53, or impart oncogenic effects. At present there is considerable discord in the field concerning these concepts with some favoring a tumor suppressor role for the p53 family members and others an oncogenic influence. In support of a tumor suppressor role is the ability of p73 and p63 isoforms to transactivate p53 target genes and the large body of work linking p73, and to some extent p63, in apoptotic events in response to cellular stresses generally considered the purview of p53. More recently, p73 has been implicated in cell death following T cell activation, the response of cancers to chemotherapy, and finally, along with p63, to the function of p53 itself. Opposing this view is the fact that the p73 and p63 genes are rarely mutated in cancers and the stark absence of tumors in the p73 null mouse. Moreover, the high expression of dominant negative (dn) versions of the p73 and p63 proteins supports an anti-p53 function and therefore possibly an oncogenic effect. Indeed, the p63 gene is located in a region of chromosome three amplified in squamous cell carcinomas and the number of reports of dn-p63 overexpression in these diseases is increasing. This review will examine both sides of these arguments in an attempt to decipher common themes and to identify opportunities these genes represent for understanding tumorigenesis.
Keyword
MeSH Terms
Figure
Reference
-
1. Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999; 399:809–813. PMID: 10391250.2. Augustin M, et al. Cloning and chromosomal mapping of the human p53-related KET gene to chromosome 3q27 and its murine homolog Ket to mouse chromosome 16. Mamm Genome. 1998; 9:899–902. PMID: 9799841.
Article3. Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, Attard M, Reelfs O, Gusterson B, Bell AK, Heath V, Tavassoli M, Farrell PJ, Smith P, Lu X, Crook T. P53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003; 3:387–402. PMID: 12726864.
Article4. Brodsky MH, et al. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000; 101:103–113. PMID: 10778860.
Article5. Di Como CJ, Gaiddon C, Prives C. P73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999; 19:1438–1449. PMID: 9891077.
Article6. Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992; 356:215–221. PMID: 1552940.
Article7. Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. P63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002; 416:560–564. PMID: 11932750.
Article8. Geddert H, Kiel S, Heep HJ, Gabbert HE, Sarbia M. The role of p63 and deltaNp63 (p40) protein expression and gene amplification in esophageal carcinogenesis. Hum Pathol. 2003; 34:850–856. PMID: 14562279.9. Gerhard J, Kirschner M. Cells, Embryos, and Evolution. 1997. Malden, MA: Blackwell Science.10. Gong JG, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999; 399:806–809. PMID: 10391249.
Article11. Heselmeyer K, et al. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc. Natl Acad Sci USA. 1996; 93:479–484.12. Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA. 2000; 97:5462–5467. PMID: 10805802.
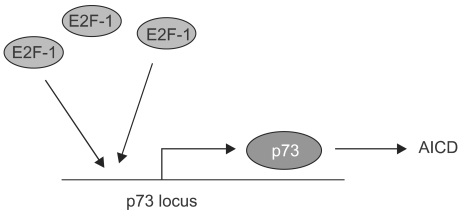
Article13. Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG Jr. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000; 407:645–648. PMID: 11034215.
Article14. Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG Jr. Chemosensitivity linked to p73 function. Cancer Cell. 2003; 3:403–410. PMID: 12726865.
Article15. Jost CA, Marin MC, Kaelin WG Jr. P73 is a simian p53-related protein that can induce apoptosis. Nature. 1997; 389:191–194. PMID: 9296498.16. Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997; 90:809–819. PMID: 9288759.
Article17. Levine A. P53, the cellular gatekeeper for growth and division. Cell. 1997; 88:323–331. PMID: 9039259.
Article18. Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000; 407:642–645. PMID: 11034214.
Article19. Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, Vousden KH, Allday MJ, Gusterson B, Ikawa S, Hinds PW, Crook T, Kaelin WG Jr. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000; 25:47–54. PMID: 10802655.
Article20. Ollmann M. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000; 101:91–101. PMID: 10778859.
Article21. Osada M, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998; 4:839–843. PMID: 9662378.
Article22. Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, Gulino A, Levrero M. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003; 5:552–558. PMID: 12766778.
Article23. Phillips AC, Vousden KH. E2F-1 induced apoptosis. Apoptosis. 2001; 6:173–182. PMID: 11388666.24. Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000; 289:304–306. PMID: 10894779.
Article25. Redon R, Muller D, Caulee K, Wanherdrick K, Abecassis J, du Manoir S. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001; 61:4122–4129. PMID: 11358835.26. Senoo M, Seki N, Ohira M, Sugano S, Watanabe M, Inuzuka S, Okamoto T, Tachibana M, Tanaka T, Shinkai Y, Kato H. A second p53-related protein, p73L, with high homology to p73. Biochem Biophys Res Commun. 1998; 30:603–607. PMID: 9703973.
Article27. Stiewe T, Putzer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000; 26:464–469. PMID: 11101847.
Article28. Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998; 4:747–748. PMID: 9662346.
Article30. Versteeg R, et al. 1p36: every subband a suppressor? Eur J Cancer. 1995; 31:538–541. PMID: 7576962.
Article31. Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. P63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998; 2:305–316. PMID: 9774969.
Article32. Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999; 398:714–718. PMID: 10227294.
Article33. Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002; 18:90–95. PMID: 11818141.
Article34. Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000; 1:199–207. PMID: 11252895.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Loss of Heterozygosity of p73, APC, and p53 in Hepatoblastoma
- p73 Methylation in Extranodal NK/T Cell Lympoma
- Expression of p73 in Non-small Cell Lung Carcinomas
- Expression of p73 in Null-p53 SKOV3 Ovarian Cancer Cell Line
- Study on mRNA expression of p21 and p73 in the cell lines of primary and metastatic squamous cell carcinoma