Cancer Res Treat.
2012 Dec;44(4):251-261.
The Blocking of c-Met Signaling Induces Apoptosis through the Increase of p53 Protein in Lung Cancer
- Affiliations
-
- 1Brain Korea 21 Project for Biomedical Science, Korea University College of Medicine, Seoul, Korea. yhk0215@korea.ac.kr
- 2Genomic Research Center for Lung and Breast/Ovarian Cancers, Korea University College of Medicine, Seoul, Korea.
- 3Laboratory of Radiation Tumor Physiology, Korea Institute of Radiological and Medical Science, Korea University College of Medicine, Seoul, Korea.
- 4Division of Oncology/Hematology, Department of Internal Medicine, Korea University College of Medicine, Seoul, Korea.
Abstract
- PURPOSE
c-Met is an attractive potential target for novel therapeutic inhibition of human cancer, and c-Met tyrosine kinase inhibitors (TKIs) are effective growth inhibitors of various malignancies. However, their mechanisms in anticancer effects are not clear. In the present study, we investigated the possibility that blocking c-Met signaling induces p53-mediated growth inhibition in lung cancer.
MATERIALS AND METHODS
The growth inhibitory effects of c-Met TKI (SU11274) on lung cancer cells and a xenograft model were assessed using the MTT assay, flow cytometry, and terminal deoxyribonucleotide transferase-mediated nick-end labeling staining. The role of p53 protein in the sensitivity of c-Met TKI (SU11274) was examined by Western blot analysis and immunohistochemistry.
RESULTS
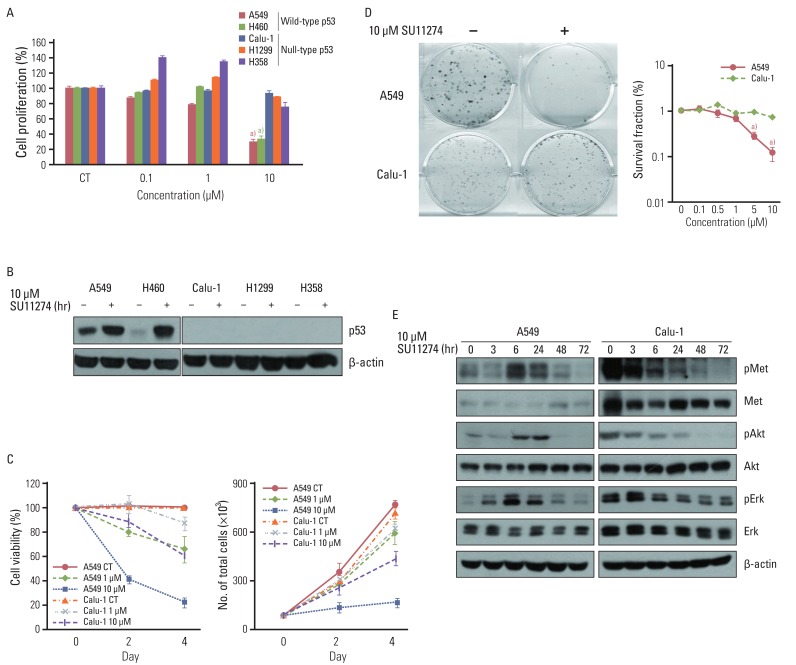
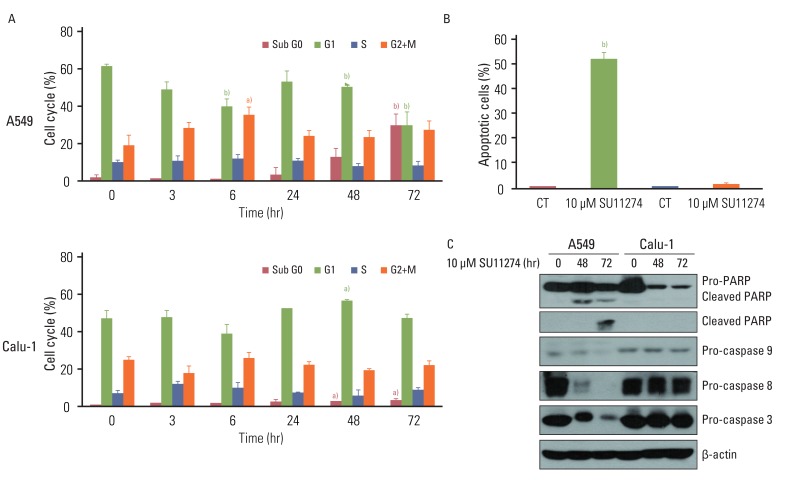
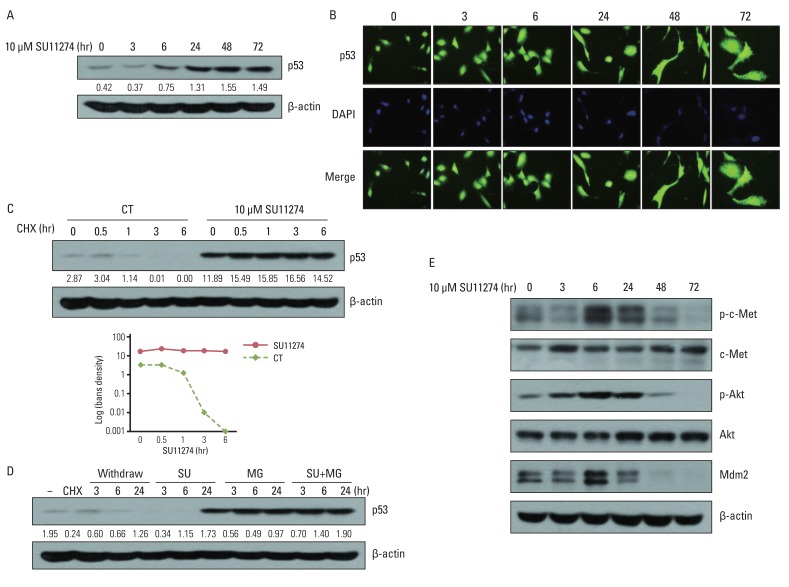
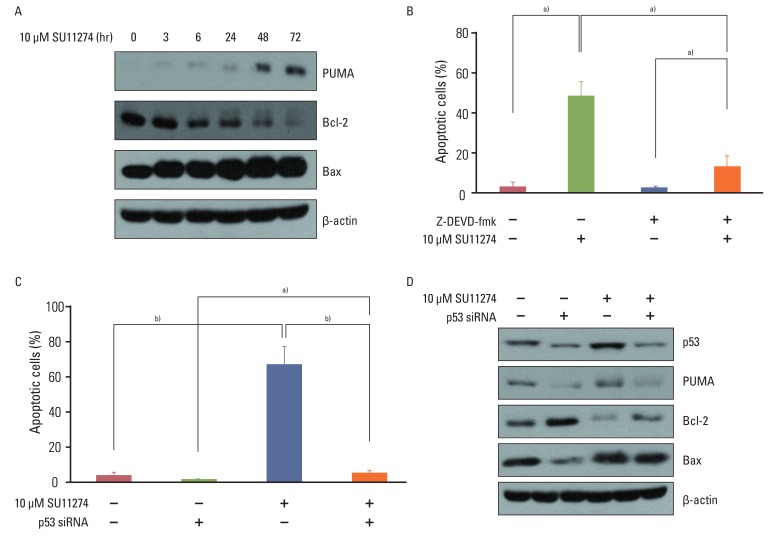
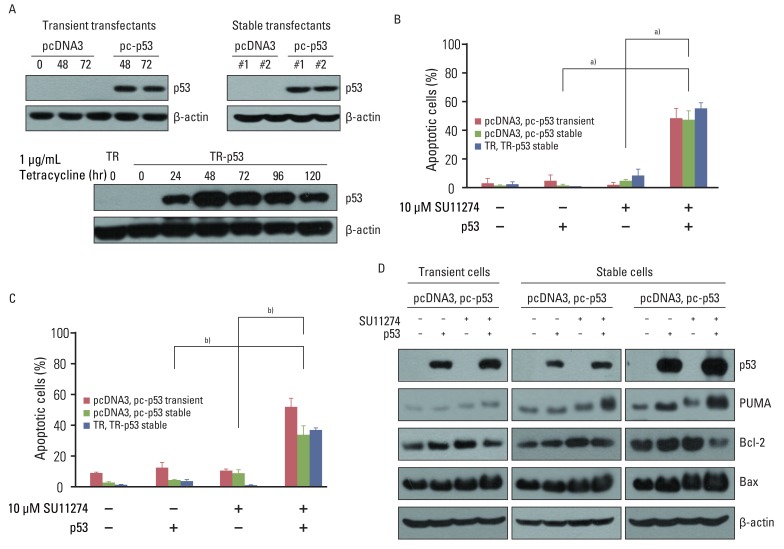
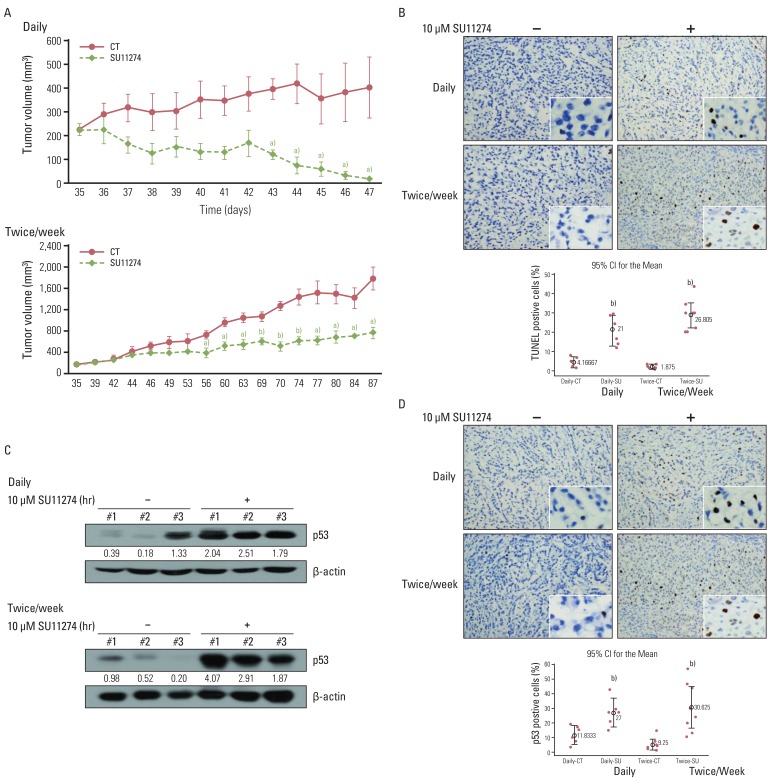
SU11274 significantly induced apoptosis in A549 cells with wild-type p53, compared with that in Calu-1 cells with null-type p53. SU11274 increased p53 protein by enhancing the stability of p53 protein. Increased p53 protein by SU11274 induced up-regulation of Bax and PUMA expression and down-regulation of Bcl-2 expression, subsequently activating caspase 3. In p53 knock-out and knock-in systems, we confirmed that SU11274 caused apoptosis through the p53-mediated apoptotic pathway. Likewise, in the A549 xenograft model, SU11274 effectively shrank tumor volume and induced apoptosis via increased p53 protein expression. Blocking c-Met signaling increased the level of p53 protein.
CONCLUSION
Our finding suggested that p53 plays an important role in SU11274-induced apoptosis, and p53 status seems to be related to the sensitivity to SU11274 in lung cancer.
Keyword
MeSH Terms
-
Apoptosis
Blotting, Western
Caspase 3
Down-Regulation
Flow Cytometry
Growth Inhibitors
Humans
Indoles
Lung
Lung Neoplasms
Molecular Targeted Therapy
Piperazines
Protein-Tyrosine Kinases
Puma
Sulfonamides
Transplantation, Heterologous
Tumor Burden
Tumor Suppressor Protein p53
Up-Regulation
Caspase 3
Growth Inhibitors
Indoles
Piperazines
Protein-Tyrosine Kinases
Sulfonamides
Tumor Suppressor Protein p53
Figure
Reference
-
1. Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991; 251:802–804. PMID: 1846706.2. Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene. 2000; 19:5582–5589. PMID: 11114738.
Article3. Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005; 225:1–26. PMID: 15922853.
Article4. Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007; 316:1039–1043. PMID: 17463250.5. Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res. 2009; 15:2207–2214. PMID: 19318488.
Article6. Ma PC, Tretiakova MS, Nallasura V, Jagadeeswaran R, Husain AN, Salgia R. Downstream signalling and specific inhibition of c-MET/HGF pathway in small cell lung cancer: implications for tumour invasion. Br J Cancer. 2007; 97:368–377. PMID: 17667909.
Article7. Puri N, Khramtsov A, Ahmed S, Nallasura V, Hetzel JT, Jagadeeswaran R, et al. A selective small molecule inhibitor of c-Met, PHA665752, inhibits tumorigenicity and angiogenesis in mouse lung cancer xenografts. Cancer Res. 2007; 67:3529–3534. PMID: 17440059.
Article8. Trotman LC, Pandolfi PP. PTEN and p53: who will get the upper hand? Cancer Cell. 2003; 3:97–99. PMID: 12620402.
Article9. Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003; 3:117–130. PMID: 12620407.
Article10. Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001; 98:11598–11603. PMID: 11504915.
Article11. Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993; 7:1126–1132. PMID: 8319905.
Article12. Moumen A, Patane S, Porras A, Dono R, Maina F. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007; 134:1443–1451. PMID: 17329361.
Article13. Rong S, Donehower LA, Hansen MF, Strong L, Tainsky M, Jeffers M, et al. Met proto-oncogene product is overexpressed in tumors of p53-deficient mice and tumors of Li-Fraumeni patients. Cancer Res. 1995; 55:1963–1970. PMID: 7728766.14. Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res. 2006; 12:3657–3660. PMID: 16778093.
Article15. Ma PC, Maulik G, Christensen J, Salgia R. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003; 22:309–325. PMID: 12884908.16. Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003; 63:7345–7355. PMID: 14612533.17. Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007; 67:2081–2088. PMID: 17332337.
Article18. Ryan KM, Phillips AC, Vousden KH. Regulation and function of the p53 tumor suppressor protein. Curr Opin Cell Biol. 2001; 13:332–337. PMID: 11343904.
Article19. Momand J, Wu HH, Dasgupta G. MDM2: master regulator of the p53 tumor suppressor protein. Gene. 2000; 242:15–29. PMID: 10721693.20. Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene. 1993; 8:307–318. PMID: 8426740.21. Seiwert TY, Jagadeeswaran R, Faoro L, Janamanchi V, Nallasura V, El Dinali M, et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 2009; 69:3021–3031. PMID: 19318576.
Article22. Zimmer Y, Vaseva AV, Medova M, Streit B, Blank-Liss W, Greiner RH, et al. Differential inhibition sensitivities of MET mutants to the small molecule inhibitor SU11274. Cancer Lett. 2010; 289:228–236. PMID: 19783361.
Article23. Xiong L, Kou F, Yang Y, Wu J. A novel role for IGF-1R in p53-mediated apoptosis through translational modulation of the p53-Mdm2 feedback loop. J Cell Biol. 2007; 178:995–1007. PMID: 17846171.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Quercetin induces cell death by caspase-dependent and p38 MAPK pathway in EGFR mutant lung cancer cells
- The Immunohistochemical analysis for the expression of survivin, an inhibitor of apoptosis protein, in non-small cell lung cancer
- p53 signaling is involved in leptin-induced growth of hepatic and breast cancer cells
- The Role of p53 in Marijuana Smoke Condensates-induced Genotoxicity and Apoptosis
- The Antitumor Effect of C-terminus of Hsp70-Interacting Protein via Degradation of c-Met in Small Cell Lung Cancer