J Korean Med Sci.
2007 Sep;22(Suppl):S24-S31. 10.3346/jkms.2007.22.S.S24.
Downregulation of the RUNX3 Gene by Promoter Hypermethylation and Hemizygous Deletion in Breast Cancer
- Affiliations
-
- 1Department of Surgery, Seoul National University Boramae Hospital, Seoul, Korea.
- 2Department of Surgery, Seoul National University College of Medicine, Seoul, Korea. dynoh@plaza.snu.ac.kr
- 3Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 1785785
- DOI: http://doi.org/10.3346/jkms.2007.22.S.S24
Abstract
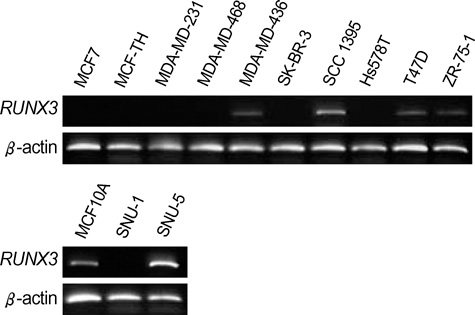
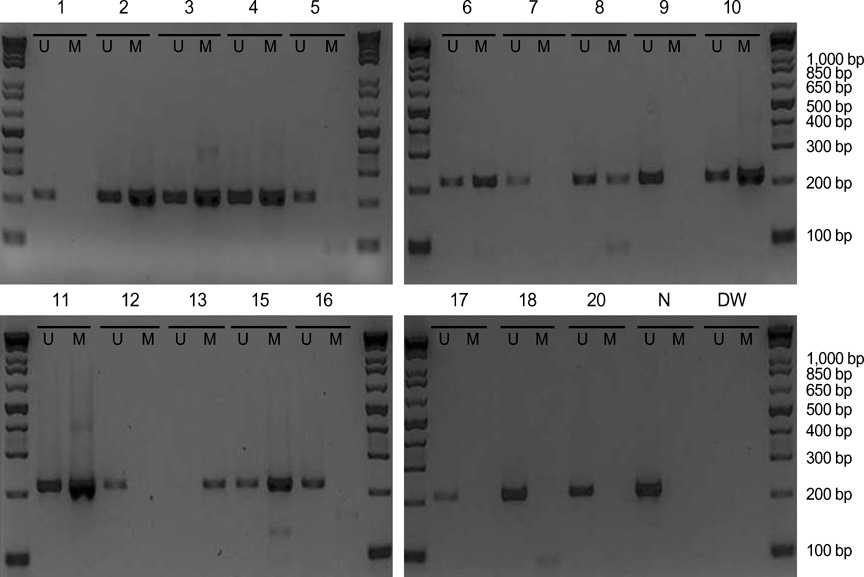
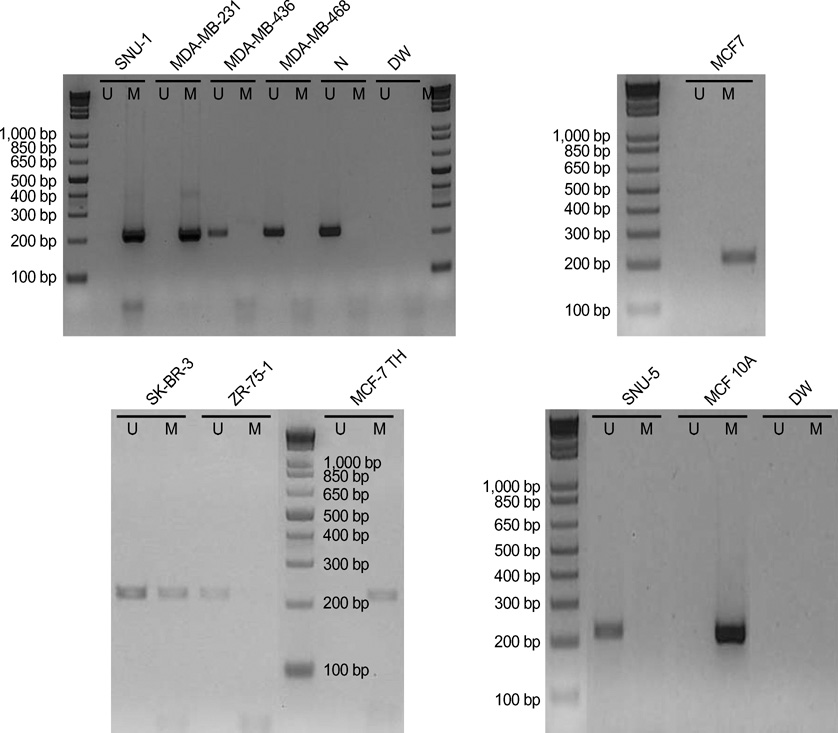
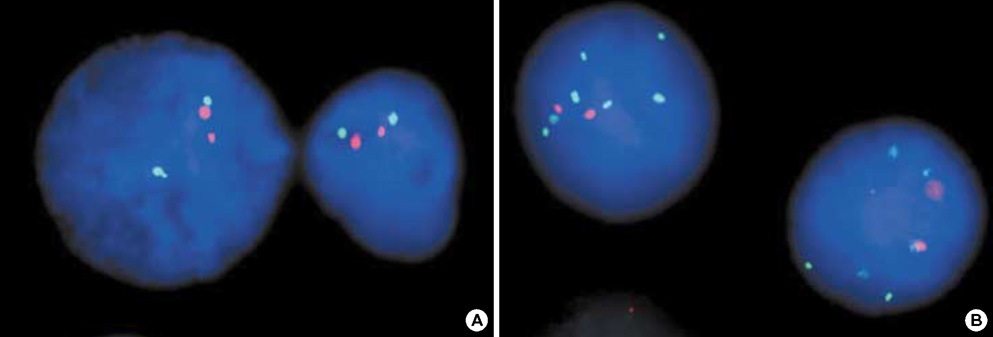
- The RUNX3 gene is regarded as a tumor suppressor gene in many human solid tumors, and its inactivation is believed to be related with solid tumor carcinogenesis. As little information is available about the role of the RUNX3 gene in breast cancer, we investigated the relationship between the RUNX3 gene and breast cancer. We performed reverse transcriptase-polymerases chain reaction (RT-PCR), methylation specific PCR, and bicolor fluorescent in situ hybridization analysis in an effort to reveal related mechanisms. Forty breast tissue samples and 13 cell lines were used in this study. Eighty-five percent of breast cancer tissues showed downregulated RUNX3 gene expression, whereas it was downregulated in only 25% of normal breast tissues by RT-PCR assay. Sixty-seven percent of breast cancer cell lines showed downregulated RUNX3 expression, but the RUNX3 gene was not expressed in two normal breast cell lines. Hypermethylation was observed in 53% of breast cancer tissues and 57% of breast cancer cell lines. Hemizygous deletion was observed in 43% of breast cancer cell lines. Hypermethylation and/or hemizygous deletion was observed in 5 of 7 breast cancer cell lines, and the four of these five examined showed no RUNX3 gene expression. We suggest that various mechanisms, including methylation and hemizygous deletion, could contribute to RUNX3 gene inactivation.
Keyword
MeSH Terms
-
Base Sequence
Breast Neoplasms/*genetics
Carcinoma, Ductal, Breast/*genetics
Case-Control Studies
Cell Line, Tumor
Core Binding Factor Alpha 3 Subunit/*genetics
DNA Methylation
DNA, Neoplasm/genetics
Down-Regulation
Female
Gene Deletion
Humans
In Situ Hybridization, Fluorescence
Promoter Regions, Genetic
Reverse Transcriptase Polymerase Chain Reaction
Figure
Cited by 1 articles
-
EMP3 Overexpression in Primary Breast Carcinomas is not Associated with Epigenetic Aberrations
Wei Zhou, Zheng Jiang, Xingang Li, Fenghua Xu, Yanbing Liu, Peie Wen, Li Kong, Ming Hou, Jinming Yu
J Korean Med Sci. 2009;24(1):97-103. doi: 10.3346/jkms.2009.24.1.97.
Reference
-
1. van Wijnen AJ, Stein GS, Gergen JP, Groner Y, Hiebert SW, Ito Y, Liu P, Neil JC, Ohki M, Speck N. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004. 23:4209–4210.
Article2. Ito Y. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells. 1999. 4:685–696.
Article3. Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int. 2003. 27:315–324.
Article4. Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005. 5:376–387.
Article5. Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002. 111:621–633.
Article6. Levanon D, Glusman G, Bangsow T, Ben-Asher E, Male DA, Avidan N, Bangsow C, Hattori M, Taylor TD, Taudien S, Blechschmidt K, Shimizu N, Rosenthal A, Sakaki Y, Lancet D, Groner Y. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001. 262:23–33.
Article7. Specchia G, Albano F, Anelli L, Zagaria A, Liso A, La Starza R, Mancini M, Sebastio L, Giugliano E, Saglio G, Liso V, Rocchi M. Insertions generating the 5'RUNX1/3'CBFA2T1 gene in acute myeloid leukemia cases show variable breakpoints. Genes Chromosomes Cancer. 2004. 41:86–91.
Article8. Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004. 23:4284–4296.
Article9. Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994. 23:425–432.
Article10. Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004. 23:4315–4329.
Article11. Zheng Q, Sebald E, Zhou G, Chen Y, Wilcox W, Lee B, Krakow D. Dysregulation of chondrogenesis in human cleidocranial dysplasia. Am J Hum Genet. 2005. 77:305–312.
Article12. Bae SC, Takahashi E, Zhang YW, Ogawa E, Shigesada K, Namba Y, Satake M, Ito Y. Cloning, mapping and expression of PEBP2 alpha C, a third gene encoding the mammalian Runt domain. Gene. 1995. 159:245–248.13. Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002. 109:113–124.
Article14. Sakakura C, Hagiwara A, Miyagawa K, Nakashima S, Yoshikawa T, Kin S, Nakase Y, Ito K, Yamagishi H, Yazumi S, Chiba T, Ito Y. Frequent downregulation of the runt domain transcription factors RUNX1, RUNX3 and their cofactor CBFB in gastric cancer. Int J Cancer. 2005. 113:221–228.15. Kim TY, Lee HJ, Hwang KS, Lee M, Kim JW, Bang YJ, Kang GH. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest. 2004. 84:479–484.
Article16. Wei D, Gong W, Oh SC, Li Q, Kim WD, Wang L, Le X, Yao J, Wu TT, Huang S, Xie K. Loss of RUNX3 expression significantly affects the clinical outcome of gastric cancer patients and its restoration causes drastic suppression of tumor growth and metastasis. Cancer Res. 2005. 65:4809–4816.
Article17. Oshimo Y, Oue N, Mitani Y, Nakayama H, Kitadai Y, Yoshida K, Ito Y, Chayama K, Yasui W. Frequent loss of RUNX3 expression by promoter hypermethylation in gastric carcinoma. Pathobiology. 2004. 71:137–143.18. Ku JL, Kang SB, Shin YK, Kang HC, Hong SH, Kim IJ, Shin JH, Han IO, Park JG. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004. 23:6736–6742.
Article19. Goel A, Arnold CN, Tassone P, Chang DK, Niedzwiecki D, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer. 2004. 112:754–759.20. Araki K, Osaki M, Nagahama Y, Hiramatsu T, Nakamura H, Ohgi S, Ito H. Expression of RUNX3 protein in human lung adenocarcinoma: implications for tumor progression and prognosis. Cancer Sci. 2005. 96:227–231.
Article21. Li QL, Kim HR, Kim WJ, Choi JK, Lee YH, Kim HM, Li LS, Kim H, Chang J, Ito Y, Youl Lee K, Bae SC. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochem Biophys Res Commun. 2004. 314:223–228.
Article22. Yanada M, Yaoi T, Shimada J, Sakakura C, Nishimura M, Ito K, Terauchi K, Nishiyama K, Itoh K, Fushiki S. Frequent hemizygous deletion at 1p36 and hypermethylation downregulate RUNX3 expression in human lung cancer cell lines. Oncol Rep. 2005. 14:817–822.
Article23. Schulmann K, Sterian A, Berki A, Yin J, Sato F, Xu Y, Olaru A, Wang S, Mori Y, Deacu E, Hamilton J, Kan T, Krasna MJ, Beer DG, Pepe MS, Abraham JM, Feng Z, Schmiegel W, Greenwald BD, Meltzer SJ. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett's-associated neoplastic progression and predicts progression risk. Oncogene. 2005. 24:4138–4148.
Article24. Mori T, Nomoto S, Koshikawa K, Fujii T, Sakai M, Nishikawa Y, Inoue S, Takeda S, Kaneko T, Nakao A. Decreased expression and frequent allelic inactivation of the RUNX3 gene at 1p36 in human hepatocellular carcinoma. Liver Int. 2005. 25:380–388.
Article25. Park WS, Cho YG, Kim CJ, Song JH, Lee YS, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY. Hypermethylation of the RUNX3 gene in hepatocellular carcinoma. Exp Mol Med. 2005. 37:276–281.
Article26. Xiao WH, Liu WW. Hemizygous deletion and hypermethylation of RUNX3 gene in hepatocellular carcinoma. World J Gastroenterol. 2004. 10:376–380.
Article27. Li J, Kleeff J, Guweidhi A, Esposito I, Berberat PO, Giese T, Buchler MW, Friess H. RUNX3 expression in primary and metastatic pancreatic cancer. J Clin Pathol. 2004. 57:294–299.28. Kato N, Tamura G, Fukase M, Shibuya H, Motoyama T. Hypermethylation of the RUNX3 gene promoter in testicular yolk sac tumor of infants. Am J Pathol. 2003. 163:387–391.
Article29. Carvalho R, Milne AN, Polak M, Corver WE, Offerhaus GJ, Weterman MA. Exclusion of RUNX3 as a tumour-suppressor gene in early-onset gastric carcinomas. Oncogene. 2005. 24:8252–8258.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Hypermethylation of the RUNX3 gene in hepatocellular carcinoma
- Epigenetic inactivation of RUNX3 in colorectal cancer
- RUNX3 Methylation Status in Colonic Carcinoma and Adenoma
- Gene Promoter Hypermethylation in Tumors and Plasma of Breast Cancer Patients
- The Role of the CDH1 Promoter Hypermethylation in the Axillary Lymph Node Metastasis and Prognosis