Relationship Between the Extent of Chromosomal Losses and the Pattern of CpG Methylation in Gastric Carcinomas
- Affiliations
-
- 1Department of Microbiology, College of Medicine, The Catholic University of Korea, Seoul, Korea. rhyumung@catholic.ac.kr
- 2Department of Clinical Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 3Department of Internal Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 1781762
- DOI: http://doi.org/10.3346/jkms.2005.20.5.790
Abstract
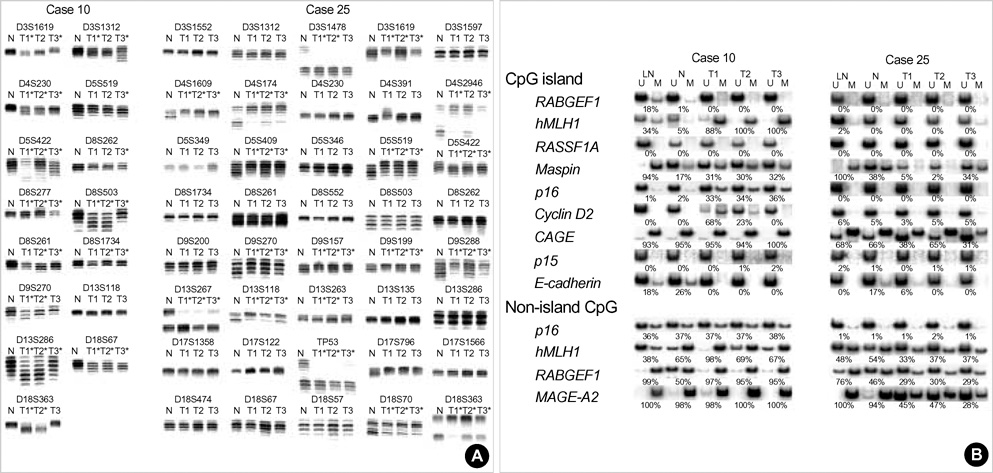
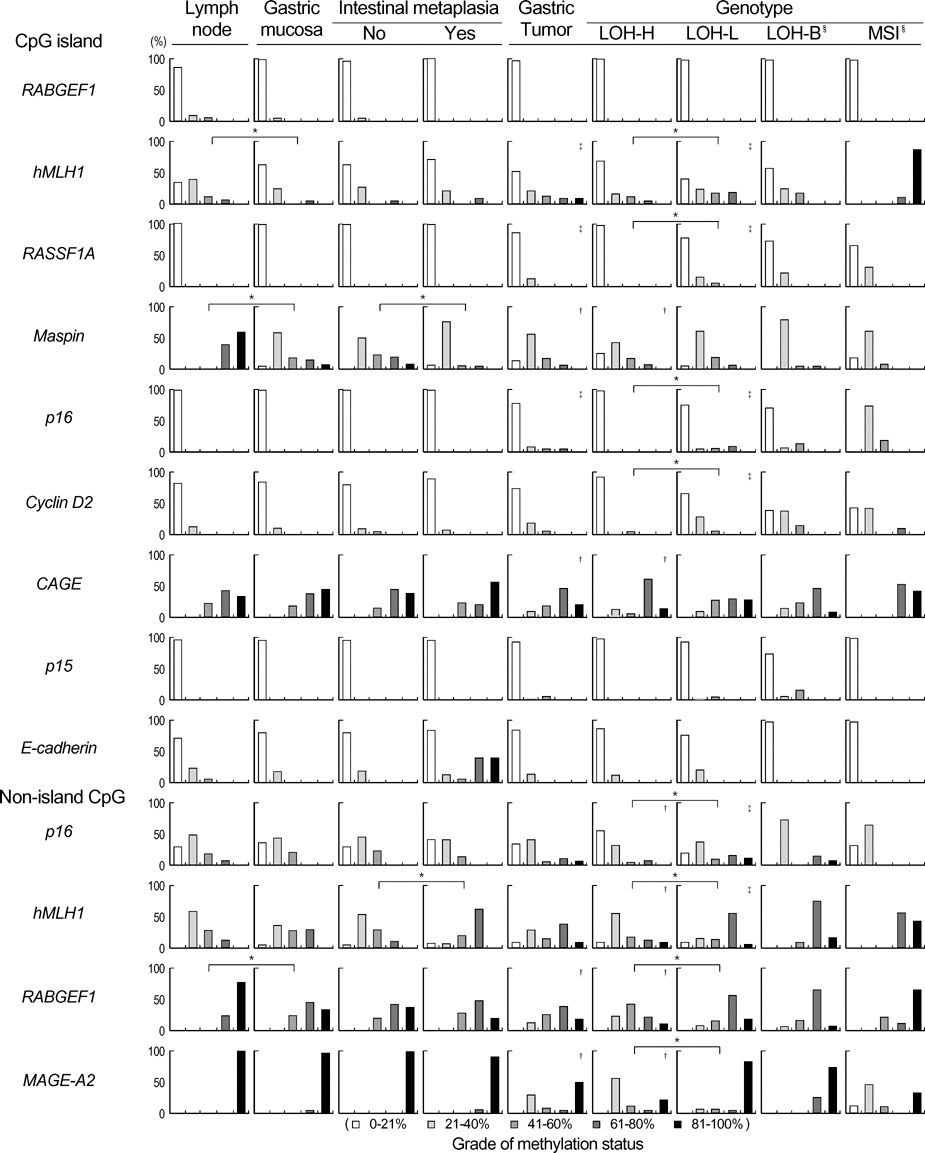
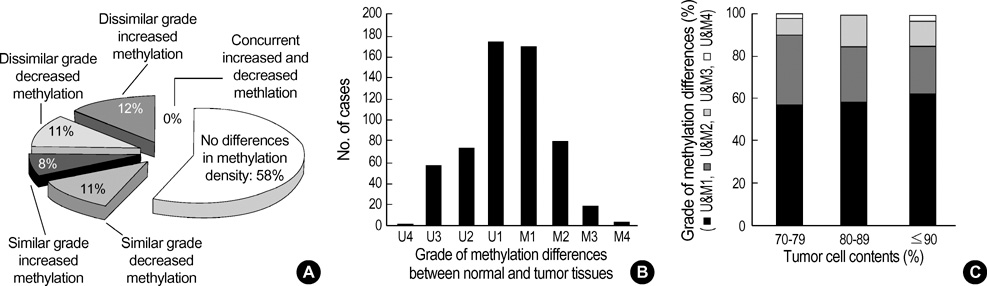
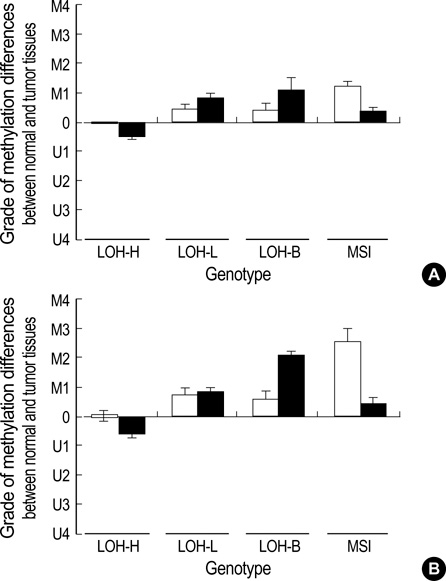
- The extent of unilateral chromosomal losses and the presence of microsatellite instability (MSI) have been classified into high-risk (high- and baseline-level loss) and low-risk (low-level loss and MSI) stem-line genotypes in gastric carcinomas. A unilateral genome-dosage reduction might stimulate compensation mechanism, which maintains the genomic dosage via CpG hypomethylation. A total of 120 tumor sites from 40 gastric carcinomas were examined by chromosomal loss analysis using 40 microsatellite markers on 8 chromosomes and methylation analysis in the 13 CpG (island/non-island) regions near the 10 genes using the bisulfite-modified DNAs. The high-level-loss tumor (four or more losses) showed a tendency toward unmethylation in the Maspin, CAGE, MAGE-A2 and RABGEF1 genes, and the other microsatellite-genotype (three or fewer losses and MSI) toward methylation in the p16, hMLH1, RASSF1A, and Cyclin D2 genes (p<0.05). The non-island CpGs of the p16 and hMLH1 genes were hypomethylated in the high-level-loss and hypermethylated in the non-high-level-loss sites (p<0.05). Consequently, hypomethylation changes were related to a high-level loss, whereas the hypermethylation changes were accompanied by a baseline-level loss, a low-level loss, or a MSI. This indicates that hypomethylation compensates the chromosomal losses in the process of tumor progression.
Keyword
MeSH Terms
-
Chromosome Aberrations/*statistics and numerical data
Chromosome Mapping/*methods
CpG Islands/*genetics
*DNA Methylation
DNA Mutational Analysis/methods
France/epidemiology
Genetic Predisposition to Disease/epidemiology/genetics
Genetic Screening/methods
Genomic Instability/genetics
Humans
Incidence
Korea/epidemiology
Microsatellite Repeats/genetics
Polymorphism, Genetic
Research Support, Non-U.S. Gov't
Risk Assessment/*methods
Risk Factors
Statistics
Stomach Neoplasms/*enzymology/*genetics
Figure
Cited by 3 articles
-
Chromosomal Losses are Associated with Hypomethylation of the Gene-Control Regions in the Stomach with a Low Number of Active Genes
Yu-Chae Jung, Seung-Jin Hong, Young-Ho Kim, Sung-Ja Kim, Seok-Jin Kang, Sang-Wook Choi, Mun-Gan Rhyu
J Korean Med Sci. 2008;23(6):1068-1089. doi: 10.3346/jkms.2008.23.6.1068.DNA Methylation and Expression Patterns of Key Tissue-specific Genes in Adult Stem Cells and Stomach Tissues
Seung-Jin Hong, Moo-Il Kang, Jung-Hwan Oh, Yu-Chae Jung, Young-Ho Kim, Sung-Ja Kim, Seung-Hye Choi, Eun-Joo Seo, Sang-Wook Choi, Mun-Gan Rhyu
J Korean Med Sci. 2009;24(5):918-929. doi: 10.3346/jkms.2009.24.5.918.DNA Methylation Patterns of Ulcer-Healing Genes Associated with the Normal Gastric Mucosa of Gastric Cancers
Seung-Jin Hong, Jung-Hwan Oh, Yu-Chae Jung, Young-Ho Kim, Sung-Ja Kim, Seok-Jin Kang, Eun-Joo Seo, Sang-Wook Choi, Moo-Il Kang, Mun-Gan Rhyu
J Korean Med Sci. 2010;25(3):405-417. doi: 10.3346/jkms.2010.25.3.405.
Reference
-
1. Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003. 33:238–244.
Article2. Lasko D, Cavenee W, Nordenskjold M. Loss of constitutional heterozygosity in human cancer. Annu Rev Genet. 1991. 25:281–314.
Article3. Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R. Allelotype of colorectal carcinomas. Science. 1989. 244:207–211.
Article4. Choi SW, Lee KJ, Bae YA, Min KO, Kwon MS, Kim KM, Rhyu MG. Genetic classification of colorectal cancer based on chromosomal loss and microsatellite instability predicts survival. Clin Cancer Res. 2002. 8:2311–2322.5. Nagel S, Borisch B, Thein SL, Oestreicher M, Nothiger F, Birrer S, Tobler A, Fey MF. Somatic mutations detected by mini- and microsatellite DNA markers reveal clonal intratumor heterogeneity in gastrointestinal cancers. Cancer Res. 1995. 55:2866–2870.6. Chung YJ, Choi JR, Park SW, Kim KM, Rhyu MG. Evidence for two modes of allelic loss: multifocal analysis on both early and advanced gastric carcinomas. Virchows Arch. 2001. 438:31–38.
Article7. Kim KM, Kwon MS, Hong SJ, Min KO, Seo EJ, Lee KY, Choi SW, Rhyu MG. Genetic classification of intestinal-type and diffuse-type gastric cancers based on chromosomal loss and microsatellite instability. Virchows Arch. 2003. 443:491–500.
Article8. Hong SJ, Choi SW, Lee KH, Lee S, Min KO, Rhyu MG. Preoperative genetic diagnosis of gastric carcinoma based on chromosomal loss and microsatellite instability. Int J Cancer. 2005. 113:249–258.
Article9. Meller VH. Dosage compensation: making 1X equal 2X. Trends Cell Biol. 2000. 10:54–59.
Article10. Muller H. Why polyploid is rarer in animals than in plants. American Nature. 1925. 59:346–353.11. Gallardo MH, Bickham JW, Honeycutt RL, Ojeda RA, Kohler N. Discovery of tetraploidy in a mammal. Nature. 1999. 401:341.
Article12. Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002. 21:5400–5413.
Article13. Lin CH, Hsieh SY, Sheen IS, Lee WC, Chen TC, Shyu WC, Liaw YF. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001. 61:4238–4243.14. Henikoff S. Heterochromatin function in complex genomes. Biochim Biophys Acta. 2000. 1470:1–8.
Article15. Lee JH, Park SJ, Abraham SC, Seo JS, Nam JH, Choi C, Juhng SW, Rashid A, Hamilton SR, Wu TT. Frequent CpG island methylation in precursor lesions and early gastric adenocarcinomas. Oncogene. 2004. 23:4646–4654.
Article16. Ward RL, Cheong K, Ku SL, Meagher A, O'Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003. 21:3729–3736.
Article17. Yang AS, Estecio MR, Garcia-Manero G, Kantarjian HM, Issa JP. Comment on "Chromosomal instability and tumors promoted by DNA hypomethylation" and "Induction of tumors in mice by genomic hypomethylation". Science. 2003. 302:1153.
Article18. Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Acta Pathol Microbiol Scand. 1965. 64:31–49.19. Greene FL, Page DL, Fleming ID, Fritz A, Balch CM, Haller DG, Morrow M. AJCC cancer staging maunal. 2002. Berlin Heidelberg New York: Springer Verlag.20. Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994. 22:2990–2997.21. Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998. 95:6870–6875.
Article22. Lo KW, Kwong J, Hui AB, Chan SY, To KF, Chan AS, Chow LS, Teo PM, Johnson PJ, Huang DP. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001. 61:3877–3881.23. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996. 93:9821–9826.
Article24. Kaneda A, Tsukamoto T, Takamura-Enya T, Watanabe N, Kaminishi M, Sugimura T, Tatematsu M, Ushijima T. Frequent hypomethylation in multiple promoter CpG islands is associated with global hypomethylation, but not with frequent promoter hypermethylation. Cancer Sci. 2004. 95:58–64.
Article25. Kokkola A, Monni O, Puolakkainen P, Nordling S, Haapiainen R, Kivilaakso E, Knuutila S. Presence of high-level DNA copy number gains in gastric carcinoma and severely dysplastic adenomas but not in moderately dysplastic adenomas. Cancer Genet Cytogenet. 1998. 107:32–36.
Article26. Akiyama Y, Maesawa C, Ogasawara S, Terashima M, Masuda T. Cell-type-specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am J Pathol. 2003. 163:1911–1919.
Article27. Cho B, Lee H, Jeong S, Bang YJ, Lee HJ, Hwang KS, Kim HY, Lee YS, Kang GH, Jeoung DI. Promoter hypomethylation of a novel cancer/testis antigen gene CAGE is correlated with its aberrant expression and is seen in premalignant stage of gastric carcinoma. Biochem Biophys Res Commun. 2003. 307:52–63.
Article28. Martin L, Assem M, Piard F. Are there several types of colorectal carcinomas? Correlations with genetic data. Eur J Cancer Prev. 1999. 8:Suppl 1. 13–20.
Article29. Bariol C, Suter C, Cheong K, Ku SL, Meagher A, Hawkins N, Ward R. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol. 2003. 162:1361–1371.
Article30. Kwon MS, Hong SJ, Cho HA, Ahn GH, Lee SS, Lee KY, Rhyu MG. Extensive and divergent chromosomal losses in squamous and spindle-cell components of esophageal sarcomatoid carcinoma. Virchows Arch. 2003. 443:635–642.
Article31. Kim KM, Kim MJ, Cho BK, Choi SW, Rhyu MG. Genetic evidence for the multi-step progression of mixed glandular-neuroendocrine gastric carcinomas. Virchows Arch. 2002. 440:85–93.
Article32. Velicescu M, Weisenberger DJ, Gonzales FA, Tsai YC, Nguyen CT, Jones PA. Cell division is required for de novo methylation of CpG islands in bladder cancer cells. Cancer Res. 2002. 62:2378–2384.33. Oue N, Oshimo Y, Nakayama H, Ito R, Yoshida K, Matsusaki K, Yasui W. DNA methylation of multiple genes in gastric carcinoma: association with histological type and CpG island methylator phenotype. Cancer Sci. 2003. 94:901–905.
Article34. Toyooka S, Toyooka KO, Harada K, Miyajima K, Makarla P, Sathyanarayana UG, Yin J, Sato F, Shivapurkar N, Meltzer SJ, Gazdar AF. Aberrant methylation of the CDH13 (H-cadherin) promoter region in colorectal cancers and adenomas. Cancer Res. 2002. 62:3382–3386.35. Song SH, Jong HS, Choi HH, Kang SH, Ryu MH, Kim NK, Kim WH, Bang YJ. Methylation of specific CpG sites in the promoter region could significantly down-regulate p16(INK4a) expression in gastric adenocarcinoma. Int J Cancer. 2000. 87:236–240.
Article36. Kang GH, Lee S, Shim YH, Kim JC, Ro JY. Profile of methylated CpG sites of hMLH1 promoter in primary gastric carcinoma with microsatellite instability. Pathol Int. 2002. 52:764–768.37. Sarbia M, Geddert H, Klump B, Kiel S, Iskender E, Gabbert HE. Hypermethylation of tumor suppressor genes (p16INK4A, p14ARF and APC) in adenocarcinomas of the upper gastrointestinal tract. Int J Cancer. 2004. 111:224–228.38. Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, Chung SC, Sung JJ, To KF. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res. 2002. 8:1761–1766.39. Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002. 161:399–403.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Chromosomal Losses are Associated with Hypomethylation of the Gene-Control Regions in the Stomach with a Low Number of Active Genes
- Methylation and Chromosomal Losses in Squamous Cell Carcinoma of the Head and Neck
- The Extent of Chromosomal Losses and the Status of CpG Methylation in Squamous Cell Carcinoma of the Head and Neck
- CpG Island Hypermethylation in Gastric Carcinoma and Its Premalignant Lesions
- DNA Methylation as Surrogate Marker For Gastric Cancer