J Korean Med Sci.
2009 Aug;24(4):571-578. 10.3346/jkms.2009.24.4.571.
Comparison of Quantitative Cytomegalovirus Real-time PCR in Whole Blood and pp65 Antigenemia Assay: Clinical Utility of CMV Real-time PCR in Hematopoietic Stem Cell Transplant Recipients
- Affiliations
-
- 1Department of Internal Medicine, The Catholic Hemopoietic Stem Cell Transplantation Center, College of Medicine, The Catholic University of Korea, Seoul, Korea. fire@catholic.ac.kr
- 2Department of Laboratory Medicine, The Catholic Hemopoietic Stem Cell Transplantation Center, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 1779186
- DOI: http://doi.org/10.3346/jkms.2009.24.4.571
Abstract
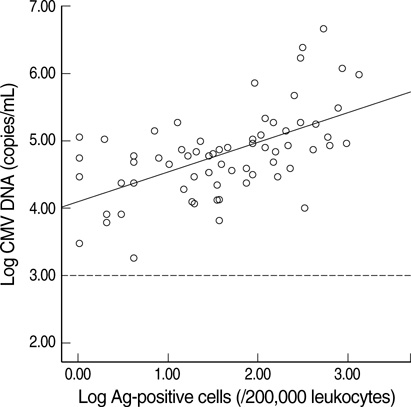
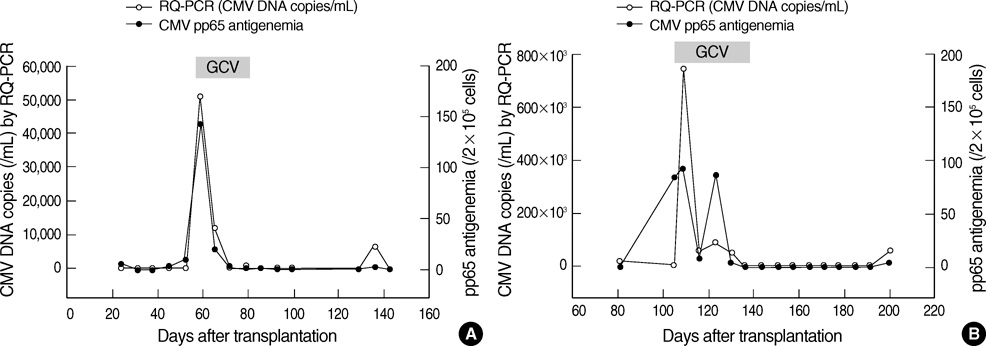
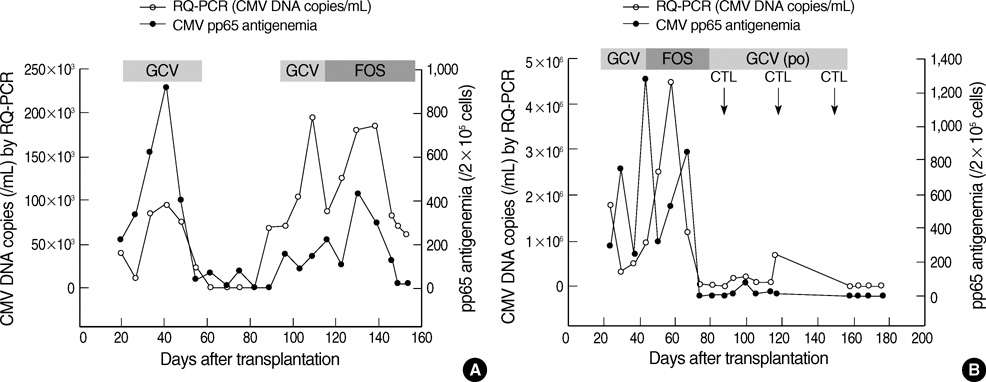
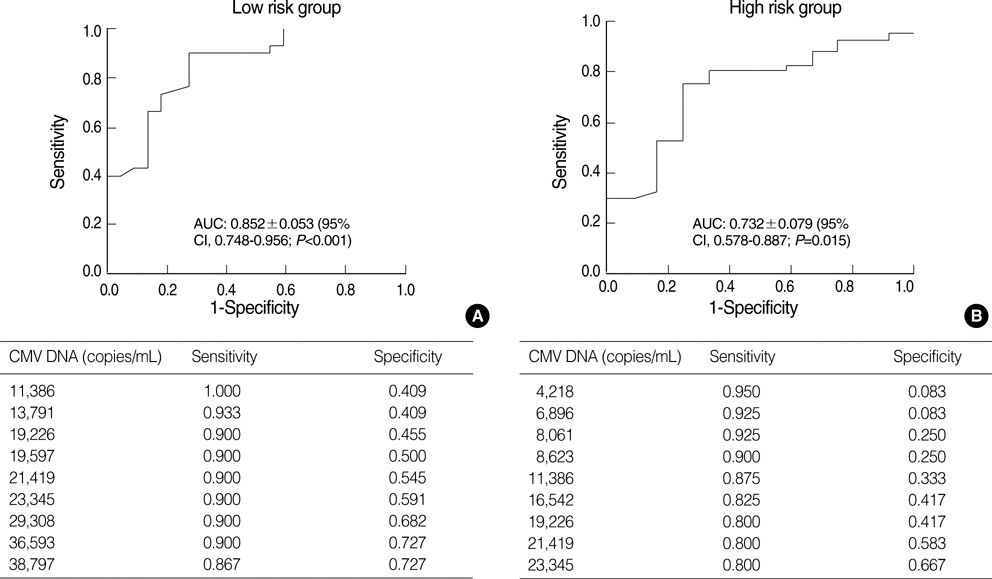
- Successful preemptive therapy for cytomegalovirus (CMV) infection in transplant patients depends on the availability of sensitive, specific, and timely diagnostic tests for CMV infection. Although the pp65 antigenemia assay has been widely used for this purpose, real-time quantification of CMV DNA has recently been recognized as an alternative diagnostic approach. However, the guidelines for antiviral therapy based on real-time quantitative polymerase chain reaction (RQ-PCR) have yet to be established. From November 2004 to March 2005, a total of 555 whole blood samples from 131 hematopoietic stem cell transplant (HSCT) recipients were prospectively collected. RQ-PCR was conducted using an Artus(R) CMV LC PCR kit (QIAGEN). Both qualitative and quantitative correlations were drawn between the two methods. Exposure to the antiviral agent influenced the results of the two assays. Additionally, the discrepancy was observed at low levels of antigenemia and CMV DNA load. Via ROC curve analysis, the tentative cutoff value for preemptive therapy was determined to be approximately 2x10(4) copies/mL (sensitivity, 80.0%; specificity, 50.0%) in the high risk patients, and approximately 3x10(4) copies/mL (sensitivity, 90.0%; specificity, 70.0%) in the patients at low risk for CMV disease. Further study to validate the optimal cutoff value for the initiation of preemptive therapy is currently underway.
Keyword
MeSH Terms
-
Adolescent
Adult
Child
Child, Preschool
Cytomegalovirus/genetics/*isolation & purification
Cytomegalovirus Infections/*diagnosis/therapy
DNA, Viral/*blood
Female
*Hematopoietic Stem Cell Transplantation
Humans
Infant
Male
Middle Aged
Phosphoproteins/analysis/immunology
Polymerase Chain Reaction/*methods
ROC Curve
Reagent Kits, Diagnostic
Sensitivity and Specificity
Viral Matrix Proteins/analysis/immunology
Figure
Cited by 2 articles
-
Comparison of Quantitation of Cytomegalovirus DNA by Real-Time PCR in Whole Blood with the Cytomegalovirus Antigenemia Assay
Seonhee Kwon, Bo Kyeung Jung, Sun-Young Ko, Chang Kyu Lee, Yunjung Cho
Ann Lab Med. 2015;35(1):99-104. doi: 10.3343/alm.2015.35.1.99.SOCS1 andSOCS3 are expressed in mononuclear cells in human cytomegalovirus viremia after allogeneic hematopoietic stem cell transplantation
Seung-Hwan Shin, Ji Yoon Lee, Tae Hyang Lee, So-Hye Park, Seung-Ah Yahng, Jae-Ho Yoon, Sung-Eun Lee, Byung-Sik Cho, Dong-Gun Lee, Yoo-Jin Kim, Seok Lee, Chang-Ki Min, Seok-Goo Cho, Dong-Wook Kim, Jong-Wook Lee, Woo-Sung Min, Chong-Won Park, Hee-Je Kim
Blood Res. 2015;50(1):40-45. doi: 10.5045/br.2015.50.1.40.
Reference
-
1. Choi SM, Lee DG, Choi JH, Yoo JH, Kim YJ, Park SH, Park SN, Min CK, Lee S, Kim HJ, Kim DW, Lee JW, Min WS, Shin WS, Kim CC. Risk-adapted preemptive therapy for cytomegalovirus disease after allogeneic stem cell transplantation: a single-center experience in Korea. Int J Hematol. 2005. 81:69–74.
Article2. Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998. 11:533–554.
Article3. Razonable RR, Paya CV, Smith TF. Role of the laboratory in diagnosis and management of cytomegalovirus infection in hematopoietic stem cell and solid-organ transplant recipients. J Clin Microbiol. 2002. 40:746–752.
Article4. Li H, Dummer JS, Estes WR, Meng S, Wright PF, Tang YW. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J Clin Microbiol. 2003. 41:187–191.
Article5. Machida U, Kami M, Fukui T, Kazuyama Y, Kinoshita M, Tanaka Y, Kanda Y, Ogawa S, Honda H, Chiba S, Mitani K, Muto Y, Osumi K, Kimura S, Hirai H. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J Clin Microbiol. 2000. 38:2536–2542.
Article6. Tanaka N, Kimura H, Iida K, Saito Y, Tsuge I, Yoshimi A, Matsuyama T, Morishima T. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J Med Virol. 2000. 60:455–462.
Article7. Griscelli F, Barrois M, Chauvin S, Lastere S, Bellet D, Bourhis JH. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J Clin Microbiol. 2001. 39:4362–4369.
Article8. Tanaka Y, Kanda Y, Kami M, Mori S, Hamaki T, Kusumi E, Miyakoshi S, Nannya Y, Chiba S, Arai Y, Mitani K, Hirai H, Mutou Y. Japan Hematology and Oncology Clinical Study Group (J-HOCS). Monitoring cytomegalovirus infection by antigenemia assay and two distinct plasma real-time PCR methods after hematopoietic stem cell transplantation. Bone Marrow Transplant. 2002. 30:315–319.
Article9. Kalpoe JS, Kroes AC, de Jong MD, Schinkel J, de Brouwer CS, Beersma MF, Claas EC. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J Clin Microbiol. 2004. 42:1498–1504.
Article10. Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J Clin Microbiol. 2004. 42:1142–1148.
Article11. Weinberg A, Schissel D, Giller R. Molecular methods for cytomegalovirus surveillance in bone marrow transplant recipients. J Clin Microbiol. 2002. 40:4203–4206.
Article12. Yoo JH, Shin WS, Choi JH, Kim DW, Lee JW, Hahn CW, Min WS, Kim CC, Kim DJ. Clinical analysis of cytomegalovirus antigenemia after allogeneic bone marrow transplantation and a suggestion of a guideline to initiation of therapy. Korean J Hematol Stem Cell Transplant. 1997. 2:43–49.13. Mengelle C, Sandres-Sauné K, Pasquier C, Rostaing L, Mansuy JM, Marty M, Da Silva I, Attal M, Massip P, Izopet J. Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay. J Clin Microbiol. 2003. 41:3840–3845.
Article14. Yerly S, Perrin L, Van Delden C, Schaffer S, Thamm S, Wunderli W, Kaiser L. Cytomegalovirus quantification in plasma by an automated real-time PCR assay. J Clin Virol. 2007. 38:298–303.
Article15. Von Müller L, Hinz J, Bommer M, Hampl W, Kluwick S, Wiedmann M, Bunjes D, Mertens T. CMV monitoring using blood cells and plasma: a comparison of apples with oranges? Bone Marrow Transplant. 2007. 39:353–357.
Article16. Razonable RR, Brown RA, Wilson J, Groettum C, Kremers W, Espy M, Smith TF, Paya CV. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation. 2002. 73:968–973.
Article17. Lilleri D, Baldanti F, Gatti M, Rovida F, Dossena L, De Grazia S, Torsellini M, Gerna G. Clinically-based determination of safe DNA-emia cutoff levels for preemptive therapy or human cytomegalovirus infections in solid organ and hematopoietic stem cell transplant recipients. J Med Virol. 2004. 73:412–418.
Article18. Verkruyse LA, Storch GA, Devine SM, Dipersio JF, Vij R. Once daily ganciclovir as initial pre-emptive therapy delayed until threshold CMV load>or=10,000 copies/ml: a safe and effective strategy for allogeneic stem cell transplant patients. Bone Marrow Transplant. 2006. 37:51–56.19. Harrington SM, Buller RS, Storch GA, Li L, Fischer SH, Murray PR, Gea-Banacloche JC. The effect of quantification standards used in real-time CMV PCR assays on guidelines for initiation of therapy in allogeneic stem cell transplant patients. Bone Marrow Transplant. 2007. 39:237–238.
Article20. Thorne LB, Civalier C, Booker J, Fan H, Gulley ML. Analytic validation of a quantitative real-time PCR assay to measure CMV viral load in whole blood. Diagn Mol Pathol. 2007. 16:73–80.
Article21. Gerna G, Sarasini A, Lilleri D, Percivalle E, Torsellini M, Baldanti F, Revello MG. In vitro model for the study of the dissociation of increasing antigenemia and decreasing DNAemia and viremia during treatment of human cytomegalovirus infection with ganciclovir in transplant recipients. J Infect Dis. 2003. 188:1639–1647.
Article22. Nichols WG, Corey L, Gooley T, Drew WL, Miner R, Huang M, Davis C, Boeckh M. Rising pp65 antigenemia during preemptive anticytomegalovirus therapy after allogeneic hematopoietic stem cell transplantation: risk factors, correlation with DNA load, and outcomes. Blood. 2001. 97:867–874.
Article23. Pérez JL, Mariscal D, Tubau F, Niubò J, Martin R. Evaluation of the CMV-vue antigenemia assay for rapid detection of cytomegalovirus in peripheral blood leukocytes. Diagn Microbiol Infect Dis. 1994. 19:15–18.
Article24. Gerna G, Revello G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992. 30:1232–1237.
Article25. Lilleri D, Gerna G, Furione M, Bernardo ME, Giorgiani G, Telli S, Baldanti F, Locatelli F. Use of a DNAemia cut-off for monitoring human cytomegalovirus infection reduces the number of preemptively treated children and young adults receiving hematopoietic stem-cell transplantation compared with qualitative pp65 antigenemia. Blood. 2007. 110:2757–2760.
Article26. Gerna G, Lilleri D, Caldera D, Furione M, Zenone Bragotti L, Alessandrino EP. Validation of a DNAemia cutoff for preemptive therapy of cytomegalovirus infection in adult hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2008. 41:873–879.
Article27. Leruez-Ville M, Ouachée M, Delarue R, Sauget AS, Blanche S, Buzyn A, Rouzioux C. Monitoring cytomegalovirus infection in adult and pediatric bone marrow transplant recipients by a real-time PCR assay performed with blood plasma. J Clin Microbiol. 2003. 41:2040–2046.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison of Real-time PCR Methods and pp65 Antigenemia Assay to Detect Cytomegalovirus Reactivation in Hematopoietic Stem Cell Transplantation
- Comparison of Quantitation of Cytomegalovirus DNA by Real-Time PCR in Whole Blood with the Cytomegalovirus Antigenemia Assay
- Evaluation of BiosewoomTM Real-Q Cytomegalovirus Quantification kit for Cytomegalovirus Viral Load Measure
- Rapid Diagnosis of HCMV Diseases by pp65 Antigenemia Assay: Comparison of pp65 Antigenemia with Polymerase Chain Reaction
- Comparison Cytomegalovirus Qualitative Assay Using Real-Time PCR and Conventional PCR