J Vet Sci.
2014 Mar;15(1):163-166. 10.4142/jvs.2014.15.1.163.
Development of a multiplex PCR assay to detect Edwardsiella tarda, Streptococcus parauberis, and Streptococcus iniae in olive flounder (Paralichthys olivaceus)
- Affiliations
-
- 1Aquatic Biotechnology Center, College of Veterinary Medicine, Gyeongsang National University, Jinju 660-701, Korea. jungts@gnu.ac.kr
- KMID: 1737623
- DOI: http://doi.org/10.4142/jvs.2014.15.1.163
Abstract
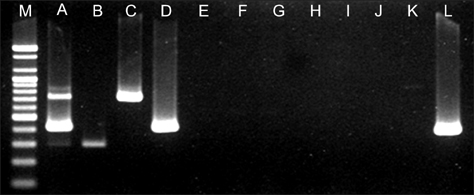
- A multiplex PCR protocol was established to simultaneously detect major bacterial pathogens in olive flounder (Paralichthys olivaceus) including Edwardsiella (E.) tarda, Streptococcus (S.) parauberis, and S. iniae. The PCR assay was able to detect 0.01 ng of E. tarda, 0.1 ng of S. parauberis, and 1 ng of S. iniae genomic DNA. Furthermore, this technique was found to have high specificity when tested with related bacterial species. This method represents a cheaper, faster, and reliable alternative for identifying major bacterial pathogens in olive flounder, the most important farmed fish in Korea.
Keyword
MeSH Terms
-
Animals
Edwardsiella tarda/genetics/*isolation & purification
Enterobacteriaceae Infections/diagnosis/microbiology/*veterinary
Fish Diseases/*diagnosis/microbiology
Fisheries/*methods
*Flatfishes
Multiplex Polymerase Chain Reaction/economics/*veterinary
Sensitivity and Specificity
Streptococcal Infections/diagnosis/microbiology/*veterinary
Streptococcus/genetics/*isolation & purification
Figure
Reference
-
1. Castro N, Toranzo AE, Núñez S, Osorio CR, Magariños B. Evaluation of four polymerase chain reaction primer pairs for the detection of Edwardsiella tarda in turbot. Dis Aquat Organ. 2010; 90:55–61.
Article2. Chang CI, Wu CC, Cheng TC, Tsai JM, Lin KJ. Multiplex nested-polymerase chain reaction for the simultaneous detection of Aeromonas hydrophila, Edwardsiella tarda, Photobacterium damselae and Streptococcus iniae, four important fish pathogens in subtropical Asia. Aquac Res. 2009; 40:1182–1190.
Article3. Chen JD, Lai SY. PCR for direct detection of Edwardsiella tarda from infected fish and environmental water by application of the hemolysin gene. Zool Stud. 1998; 37:169–176.4. Evans JJ, Klesius PH, Plumb JA, Shoemaker CA. Edwardsiella septicaemias. In : Woo PTK, Bruno DW, editors. Fish Diseases and Disorders. Vol. 3:2nd ed. wallingford: CAB International;2011. p. 512–555.5. Lan J, Zhang XH, Wang Y, Chen J, Han Y. Isolation of an unusual strain of Edwardsiella tarda from turbot and establish a PCR detection technique with the gyrB gene. J Appl Microbiol. 2008; 105:644–651.
Article6. Mata AI, Gibello A, Casamayor A, Blanco MM, Domínguez L, Fernández-Garayzábal JF. Multiplex PCR assay for detection of bacterial pathogens associated with warm-water streptococcosis in fish. Appl Environ Microbiol. 2004; 70:3183–3187.
Article7. Nho SW, Shin GW, Park SB, Jang HB, Cha IS, Ha MA, Kim YR, Park YK, Dalvi RS, Kang BJ, Joh SJ, Jung TS. Phenotypic characteristics of Streptococcus iniae and Streptococcus parauberis isolated from olive flounder (Paralichthys olivaceus). FEMS Microbiol Lett. 2009; 293:20–27.
Article8. Park SB, Aoki T, Jung TS. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet Res. 2012; 43:67.9. Sakai T, Iida T, Osatomi K, Kanai K. Detection of type 1 fimbrial genes in fish pathogenic and non-pathogenic Edwardsiella tarda strains by PCR. Fish Pathol. 2007; 42:115–117.
Article10. Sakai T, Yuasa K, Sano M, Iida T. Identification of Edwardsiella ictaluri and E. tarda by species-specific polymerase chain reaction targeted to the upstream region of the fimbrial gene. J Aquat Anim Health. 2009; 21:124–132.
Article11. Shin GW, Palaksha KJ, Yang HH, Shin YS, Kim YR, Lee EY, Kim HY, Kim YJ, Oh MJ, Yoshida T, Jung TS. Discrimination of streptococcosis agents in olive flounder (Paralichthys olivaceus). Bull Eur Assoc Fish Pathol. 2006; 26:68–79.12. Zlotkin A, Eldar A, Ghittino C, Bercovier H. Identification of Lactococcus garvieae by PCR. J Clin Microbiol. 1998; 36:983–985.13. Cho MY, Oh YK, Lee DC, Kim JH, Park MA. Geographical comparison on different methods for identification of Streptococcus parauberis isolated from cultured olive flounder, Paralichthys olivaceus. J Fish Pathol. 2007; 20:49–60.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Isolation and characterization of Streptococcus sp. from diseased flounder (Paralichthys olivaceus) in Jeju Island
- Kudoa ogawai (Myxosporea: Kudoidae) Infection in Cultured Olive Flounder Paralichthys olivaceus
- Antimicrobial activity of essential oil of Eucalyptus globulus against fish pathogenic bacteria
- Gliotoxin is Antibacterial to Drug-resistant Piscine Pathogens
- Sinuolinea capsularis (Myxosporea: Sinuolineidae) Isolated from Urinary Bladder of Cultured Olive Flounder Paralichthys olivaceus