Brain Tumor Res Treat.
2014 Apr;2(1):7-21. 10.14791/btrt.2014.2.1.7.
Altered Histone Modifications in Gliomas
- Affiliations
-
- 1Division of Neuro-Oncology, Department of Neurosurgery, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, Changwon, Korea. yzkim@skku.edu
- KMID: 1734663
- DOI: http://doi.org/10.14791/btrt.2014.2.1.7
Abstract
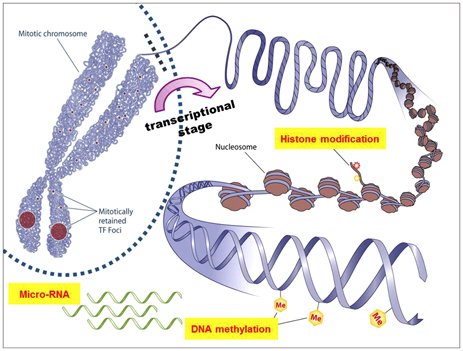
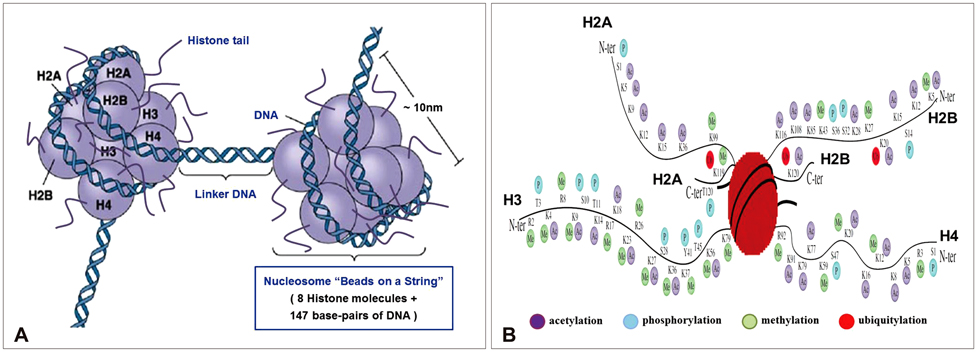
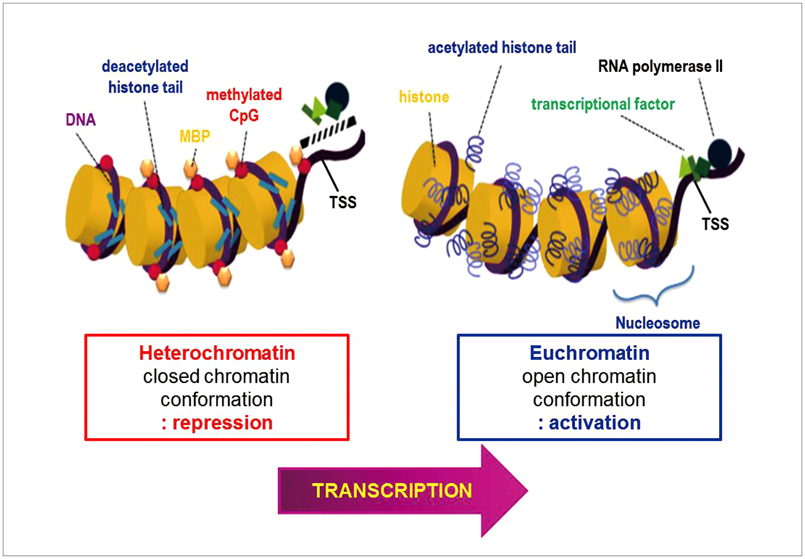
- Gliomas are the most frequently occurring primary brain tumors in adults. Although they exist in different malignant stages, including histologically benign forms and highly aggressive states, most gliomas are clinically challenging for neuro-oncologists because of their infiltrative growth patterns and inherent relapse tendency with increased malignancy. Once this disease reaches the glioblastoma multiforme stage, the prognosis of patients is dismal: median survival time is 15 months. Extensive genetic analyses of glial tumors have revealed a variety of deregulated genetic pathways involved in DNA repair, apoptosis, cell migration/adhesion, and cell cycle. Recently, it has become evident that epigenetic alterations may also be an important factor for glioma genesis. Of epigenetic marks, histone modification is a key mark that regulates gene expression and thus modulates a wide range of cellular processes. In this review, I discuss the neuro-oncological significance of altered histone modifications and modifiers in glioma patients while briefly overviewing the biological roles of histone modifications.
Keyword
MeSH Terms
Figure
Reference
-
1. Clark SJ, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995; 10:20–27.
Article2. Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998; 19:219–220.
Article3. Hansen RS, Gartler SM. 5-Azacytidine-induced reactivation of the human X chromosome-linked PGK1 gene is associated with a large region of cytosine demethylation in the 5' CpG island. Proc Natl Acad Sci U S A. 1990; 87:4174–4178.
Article4. Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993; 366:362–365.
Article5. Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003; 300:455.
Article6. Fouse SD, Shen Y, Pellegrini M, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008; 2:160–169.
Article7. Turner BM. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat Struct Mol Biol. 2005; 12:110–112.
Article8. Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007; 76:75–100.
Article9. Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999; 98:285–294.
Article10. Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004; 14:R546–R551.
Article11. Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005; 21:133–153.
Article12. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705.
Article13. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007; 8:286–298.
Article14. Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005; 37:391–400.
Article15. Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964; 51:786–794.
Article16. Loidl P. Histone acetylation: facts and questions. Chromosoma. 1994; 103:441–449.
Article17. Walkinshaw DR, Tahmasebi S, Bertos NR, Yang XJ. Histone deacetylases as transducers and targets of nuclear signaling. J Cell Biochem. 2008; 104:1541–1552.
Article18. Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007; 8:284–295.
Article19. Ekwall K. Genome-wide analysis of HDAC function. Trends Genet. 2005; 21:608–615.
Article20. Steger DJ, Workman JL. Remodeling chromatin structures for transcription: what happens to the histones? Bioessays. 1996; 18:875–884.
Article21. Fletcher TM, Hansen JC. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J Biol Chem. 1995; 270:25359–25362.
Article22. Norton VG, Imai BS, Yau P, Bradbury EM. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989; 57:449–457.
Article23. Clarke S. Protein methylation. Curr Opin Cell Biol. 1993; 5:977–983.
Article24. Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry. 1964; 3:10–15.25. Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998; 61:65–131.
Article26. Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat Struct Biol. 2000; 7:1165–1171.27. Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004; 24:9630–9645.
Article28. Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002; 12:1052–1058.
Article29. Sims RJ 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003; 19:629–639.
Article30. Santos-Rosa H, Kirmizis A, Nelson C, et al. Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol. 2009; 16:17–22.
Article31. Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004; 119:941–953.
Article32. Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006; 439:811–816.
Article33. Anand R, Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem. 2007; 282:35425–35429.
Article34. Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006; 125:467–481.
Article35. Jenuwein T, Allis CD. Translating the histone code. Science. 2001; 293:1074–1080.
Article36. Rea S, Eisenhaber F, O'Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000; 406:593–599.
Article37. Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005; 438:1176–1180.
Article38. Metzger E, Imhof A, Patel D, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010; 464:792–796.
Article39. Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002; 418:104–108.
Article40. Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001; 410:120–124.
Article41. Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998; 67:545–579.
Article42. Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997; 22:197–202.
Article43. Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000; 408:495–498.
Article44. Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006; 75:243–269.
Article45. Sims RJ 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006; 20:2779–2786.
Article46. Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005; 437:436–439.
Article47. Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003; 194:77–95.
Article48. van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005; 6:757–765.
Article49. Shroff R, Arbel-Eden A, Pilch D, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004; 14:1703–1711.
Article50. Downs JA, Allard S, Jobin-Robitaille O, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004; 16:979–990.
Article51. Downs JA, Côté J. Dynamics of chromatin during the repair of DNA double-strand breaks. Cell Cycle. 2005; 4:1373–1376.
Article52. Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005; 436:294–298.
Article53. Murr R, Vaissière T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007; 26:5358–5372.
Article54. Botuyan MV, Lee J, Ward IM, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006; 127:1361–1373.
Article55. Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004; 119:603–614.
Article56. Sun Y, Jiang X, Xu Y, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009; 11:1376–1382.
Article57. Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002; 420:198–202.
Article58. Cimbora DM, Schübeler D, Reik A, et al. Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol Cell Biol. 2000; 20:5581–5591.
Article59. Burke TW, Cook JG, Asano M, Nevins JR. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem. 2001; 276:15397–15408.
Article60. Doyon Y, Cayrou C, Ullah M, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006; 21:51–64.
Article61. Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010; 17:430–437.
Article62. Jung KW, Ha J, Lee SH, Won YJ, Yoo H. An updated nationwide epidemiology of primary brain tumors in republic of Korea. Brain Tumor Res Treat. 2013; 1:16–23.
Article63. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008; 359:492–507.
Article64. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008; 321:1807–1812.
Article65. Campos B, Bermejo JL, Han L, et al. Expression of nuclear receptor corepressors and class I histone deacetylases in astrocytic gliomas. Cancer Sci. 2011; 102:387–392.
Article66. Schmidt N, Windmann S, Reifenberger G, Riemenschneider MJ. DNA hypermethylation and histone modifications downregulate the candidate tumor suppressor gene RRP22 on 22q12 in human gliomas. Brain Pathol. 2012; 22:17–25.
Article67. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326.
Article68. Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007; 39:237–242.
Article69. Di Croce L. Chromatin modifying activity of leukaemia associated fusion proteins. Hum Mol Genet. 2005; 14(Spec No 1):R77–R84.
Article70. Petrij F, Giles RH, Dauwerse HG, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995; 376:348–351.
Article71. Häyry V, Tanner M, Blom T, et al. Copy number alterations of the polycomb gene BMI1 in gliomas. Acta Neuropathol. 2008; 116:97–102.
Article72. Lucio-Eterovic AK, Cortez MA, Valera ET, et al. Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer. 2008; 8:243.
Article73. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008; 321:1807–1812.
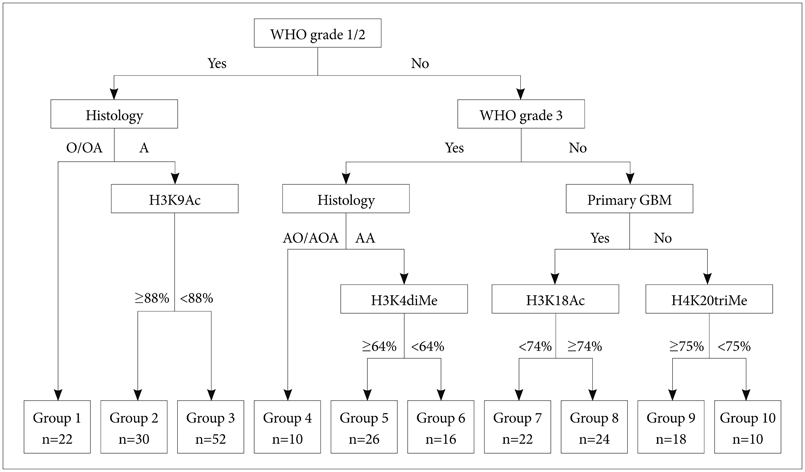
Article74. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482:226–231.75. Liu BL, Cheng JX, Zhang X, et al. Global histone modification patterns as prognostic markers to classify glioma patients. Cancer Epidemiol Biomarkers Prev. 2010; 19:2888–2896.
Article76. Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007; 5:981–989.
Article77. Sathornsumetee S, Reardon DA, Desjardins A, Quinn JA, Vredenburgh JJ, Rich JN. Molecularly targeted therapy for malignant glioma. Cancer. 2007; 110:13–24.
Article78. Yin D, Ong JM, Hu J, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor: effects on gene expression and growth of glioma cells in vitro and in vivo. Clin Cancer Res. 2007; 13:1045–1052.
Article79. Qiu L, Kelso MJ, Hansen C, West ML, Fairlie DP, Parsons PG. Anti-tumour activity in vitro and in vivo of selective differentiating agents containing hydroxamate. Br J Cancer. 1999; 80:1252–1258.
Article80. Lee JH, Park JH, Jung Y, et al. Histone deacetylase inhibitor enhances 5-fluorouracil cytotoxicity by down-regulating thymidylate synthase in human cancer cells. Mol Cancer Ther. 2006; 5:3085–3095.
Article81. Chinnaiyan P, Cerna D, Burgan WE, et al. Postradiation sensitization of the histone deacetylase inhibitor valproic acid. Clin Cancer Res. 2008; 14:5410–5415.
Article82. Entin-Meer M, Yang X, VandenBerg SR, et al. In vivo efficacy of a novel histone deacetylase inhibitor in combination with radiation for the treatment of gliomas. Neuro Oncol. 2007; 9:82–88.
Article83. Ellis L, Pan Y, Smyth GK, et al. Histone deacetylase inhibitor panobinostat induces clinical responses with associated alterations in gene expression profiles in cutaneous T-cell lymphoma. Clin Cancer Res. 2008; 14:4500–4510.
Article84. Gimsing P, Hansen M, Knudsen LM, et al. A phase I clinical trial of the histone deacetylase inhibitor belinostat in patients with advanced hematological neoplasia. Eur J Haematol. 2008; 81:170–176.
Article85. Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008; 6:21–30.
Article86. Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007; 21:1050–1063.
Article87. Zaidi SK, Young DW, Montecino M, et al. Architectural epigenetics: mitotic retention of mammalian transcriptional regulatory information. Mol Cell Biol. 2010; 30:4758–4766.
Article89. Mercurio C, Plyte S, Minucci S. Alterations of histone modifications in cancer. In : Tollefsbol T, editor. Epigenetics in human disease. Waltham: Elsevier Inc.;2012. p. 53–88.90. Hatzimichael E, Crook T. Cancer epigenetics: new therapies and new challenges. J Drug Deliv. 2013; 2013:529312.
Article91. Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010; 70:57–85.
Article92. Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP, Citterio E. The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell Div. 2008; 3:8.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Epigenetic Cross-Talk between DNA Methylation and Histone Modifications in Human Cancers
- The Role of Histone Acetylation in Mesenchymal Stem Cell Differentiation
- Epigenetic Modifications: Novel Therapeutic Approach for Thyroid Cancer
- Characterization of Chromatin Structure-associated Histone Modifications in Breast Cancer Cells
- Writing, erasing and reading histone lysine methylations