J Vet Sci.
2013 Mar;14(1):45-51. 10.4142/jvs.2013.14.1.45.
Trends in genotype frequency resulting from breeding for resistance to classical scrapie in Belgium (2006~2011)
- Affiliations
-
- 1Pathology and Prionology, Veterinary and Agrochemical Research Centre (CODA-CERVA), 1180 Brussels, Belgium. stefan.roels@coda-cerva.be
- KMID: 1482812
- DOI: http://doi.org/10.4142/jvs.2013.14.1.45
Abstract
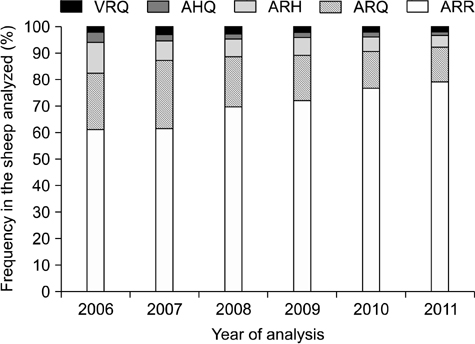
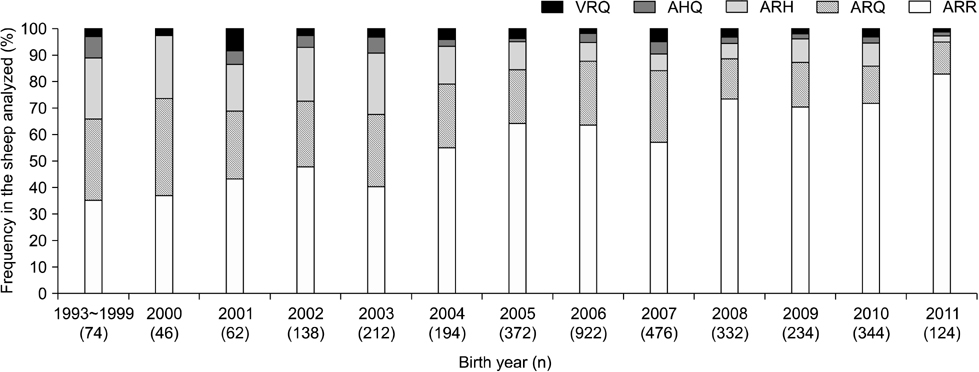
- In sheep, susceptibility to scrapie is mainly determined by codons 136, 154, and 171 of the PRNP gene. Five haplotypes are usually present (ARR, ARQ, ARH, AHQ, and VRQ). The ARR haplotype confers the greatest resistance to classical scrapie while VRQ renders animals most susceptible. In 2004, the European Union implemented a breeding program that promotes selection of the ARR haplotype while reducing the incidence of VRQ. From 2006 to 2011 in Belgium, frequency for the ARR/ARR genotypes increased from 38.3% to 63.8% (n = 6,437), the ARQ haplotype diminished from 21.1% to 12.9%, and the VRQ haplotype decreased from 2.0% to 1.7%. The status of codon 141, a determinant for atypical scrapie, was also evaluated. Out of 27 different breeds (n = 5,163), nine were abundant. The ARR/ARR frequency increased in eight of these nine major breeds. The selection program has had a major impact on the ARR haplotype frequency in Belgium. However, the occurrence of atypical scrapie represents a critical point for this program that warrants the continuous monitoring of scrapie. Additionally, genotype frequencies among the breeds varied greatly. Texel, a breed that is common in Belgium, can still be selected for due to its average ARR frequency.
MeSH Terms
Figure
Reference
-
1. Baylis M, Chihota C, Stevenson E, Goldmann W, Smith A, Sivam K, Tongue S, Gravenor MB. Risk of scrapie in British sheep of different prion protein genotype. J Gen Virol. 2004. 85(Pt 9):2735–2740.
Article2. Baylis M, Goldmann W. The genetics of scrapie in sheep and goats. Curr Mol Med. 2004. 4:385–396.
Article3. Belt PBGM, Muileman IH, Schreuder BEC, Bos-de Ruijter J, Gielkens ALJ, Smits MA. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol. 1995. 76(Pt 3):509–517.
Article4. Benestad SL, Arsac JN, Goldmann W, Nöremark M. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet Res. 2008. 39:19.
Article5. Benestad SL, Sarradin P, Thu B, Schönheit J, Tranulis MA, Bratberg B. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet Rec. 2003. 153:202–208.
Article6. Buschmann A, Biacabe AG, Ziegler U, Bencsik A, Madec JY, Erhardt G, Lühken G, Baron T, Groschup MH. Atypical scrapie cases in Germany and France are identified by discrepant reaction patterns in BSE rapid tests. J Virol Methods. 2004. 117:27–36.
Article7. De Bosschere H, Roels S, Benestad SL, Vanopdenbosch E. Scrapie case similar to Nor98 diagnosed in Belgium via active surveillance. Vet Rec. 2004. 155:707–708.
Article8. De Bosschere H, Roels S, Dechamps P, Vanopdenbosch E. TSE detected in a Belgian ARR-homozygous sheep via active surveillance. Vet J. 2007. 173:449–451.
Article9. Dickinson AG, Stamp JT, Renwick CC. Maternal and lateral transmission of scrapie in sheep. J Comp Pathol. 1974. 84:19–25.
Article10. Drögemüller C, Leeb T, Distl O. PrP genotype frequencies in German breeding sheep and the potential to breed for resistance to scrapie. Vet Rec. 2001. 149:349–352.
Article11. Eglin RD, Warner R, Gubbins S, Sivam SK, Dawson M. Frequencies of PrP genotypes in 38 breeds of sheep sampled in the National Scrapie Plan for Great Britain. Vet Rec. 2005. 156:433–437.
Article12. Elsen JM, Amigues Y, Schelcher F, Ducrocq V, Andreoletti O, Eychenne F, Khang JV, Poivey JP, Lantier F, Laplanche JL. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Arch Virol. 1999. 144:431–445.
Article13. European Food Safety Authority. Opinion of the Scientific Panel on Biological Hazards on "the breeding programme for TSE resistance in sheep". EFSA J. 2006. 382:1–46.14. Evoniuk JM, Berg PT, Johnson ML, Larson DM, Maddock TD, Stoltenow CL, Schauer CS, O'Rourke KI, Redmer DA. Associations between genotypes at codon 171 and 136 of the prion protein gene and production traits in market lambs. Am J Vet Res. 2007. 68:1073–1078.
Article15. Fediaevsky A, Tongue SC, Nöremark M, Calavas D, Ru G, Hopp P. A descriptive study of the prevalence of atypical and classical scrapie in sheep in 20 European countries. BMC Vet Res. 2008. 4:19.
Article16. Fryer HR, Baylis M, Sivam K, McLean AR. Quantifying the risk from ovine BSE and the impact of control strategies. Proc Biol Sci. 2007. 274:1497–1503.
Article17. Georgsson G, Sigurdarson S, Brown P. Infectious agent of sheep scrapie may persist in the environment for at least 16 years. J Gen Virol. 2006. 87(Pt 12):3737–3740.
Article18. Goldmann W. PrP genetics in ruminant transmissible spongiform encephalopathies. Vet Res. 2008. 39:30.19. Gubbins S, McIntyre KM. Prevalence of sheep infected with classical scrapie in Great Britain, 1993-2007. Epidemiol Infect. 2009. 137:787–791.
Article20. Hagenaars TJ, Melchior MB, Bossers A, Davidse A, Engel B, van Zijderveld FG. Scrapie prevalence in sheep of susceptible genotype is declining in a population subject to breeding for resistance. BMC Vet Res. 2010. 6:25.
Article21. Häusermann C, Schwermer H, Oevermann A, Nentwig A, Zurbriggen A, Heim D, Seuberlich T. Surveillance and simulation of bovine spongiform encephalopathy and scrapie in small ruminants in Switzerland. BMC Vet Res. 2010. 6:20.
Article22. Hickford JGH, Zhou H, Fang Q, Byun SO, Gong H. Frequency of PRNP genotypes in common New Zealand sheep breeds. Vet Rec. 2008. 163:453–454.23. Hunter N, Foster JD, Goldmann W, Stear MJ, Hope J, Bostock C. Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Arch Virol. 1996. 141:809–824.
Article24. Hunter N, Goldmann W, Benson G, Foster JD, Hope J. Swaledale sheep affected by natural scrapie differ significantly in PrP genotype frequencies from healthy sheep and those selected for reduced incidence of scrapie. J Gen Virol. 1993. 74(Pt 6):1025–1031.
Article25. Ianella P, McManus CM, Caetano AR, Paiva SR. PRNP haplotype and genotype frequencies in Brazilian sheep: issues for conservation and breeding programs. Res Vet Sci. 2012. 93:219–225.
Article26. Jeffrey M, González L. Classical sheep transmissible spongiform encephalopathies: pathogenesis, pathological phenotypes and clinical disease. Neuropathol Appl Neurobiol. 2007. 33:373–394.
Article27. L'Homme Y, Leboeuf A, Cameron J. PrP genotype frequencies of Quebec sheep breeds determined by real-time PCR and molecular beacons. Can J Vet Res. 2008. 72:320–324.28. Lühken G, Buschmann A, Brandt H, Eiden M, Groschup MH, Erhardt G. Epidemiological and genetical differences between classical and atypical scrapie cases. Vet Res. 2007. 38:65–80.
Article29. McIntyre KM, Trewby H, Gubbins S, Baylis M. The impact of sheep breed on the risk of classical scrapie. Epidemiol Infect. 2010. 138:384–392.
Article30. Melchior MB, Windig JJ, Hagenaars TJ, Bossers A, Davidse A, van Zijderveld FG. Eradication of scrapie with selective breeding: are we nearly there? BMC Vet Res. 2010. 6:24.
Article31. Moreno CR, Moazami-Goudarzi K, Laurent P, Cazeau G, Andreoletti O, Chadi S, Elsen JM, Calavas D. Which PrP haplotypes in a French sheep population are the most susceptible to atypical scrapie? Arch Virol. 2007. 152:1229–1232.
Article32. Moum T, Olsaker I, Hopp P, Moldal T, Valheim M, Moum T, Benestad SL. Polymorphisms at codons 141 and 154 in the ovine prion protein gene are associated with scrapie Nor98 cases. J Gen Virol. 2005. 86(Pt 1):231–235.
Article33. Prusiner SB. Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci. 1996. 21:482–487.
Article34. Roels S, Renard C, De Bosschere H, Geeroms R, Van Poucke M, Peelman L, Vanopdenbosch E. Detection of polymorphisms in the prion protein gene in the Belgian sheep population: some preliminary data. Vet Q. 2004. 26:3–11.
Article35. Saunders GC, Cawthraw S, Mountjoy SJ, Hope J, Windl O. PrP genotypes of atypical scrapie cases in Great Britain. J Gen Virol. 2006. 87(Pt 11):3141–3149.
Article36. Sild E, Volskiene R, Viinalass H, Miceikiene I, Grislis Z, Distl O, Drögemüller C. Detection of prion protein gene polymorphisms in Baltic breeds of sheep. Vet Rec. 2006. 159:247–250.
Article37. Slate J. Scrapie genetics before the discovery of prions. Heredity (Edinb). 2010. 104:122–123.
Article38. Smits MA, Bossers A, Schreuder BEC. Prion protein and scrapie susceptibility. Vet Q. 1997. 19:101–105.
Article39. Spiropoulos J, Lockey R, Sallis RE, Terry LA, Thorne L, Holder TM, Beck KE, Simmons MM. Isolation of prion with BSE properties from farmed goat. Emerg Infect Dis. 2011. 17:2253–2261.
Article40. Sweeney T, Hanrahan JP. The evidence of associations between prion protein genotype and production, reproduction, and health traits in sheep. Vet Res. 2008. 39:28.
Article41. The European Commission. Cumulative TSE Testing in Sheep Since 2002. 2009. Brussels: The European Commission;1.42. The European Commission. The TSE Road Map 2. 2010. Brussels: The European Commission;4–15.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Antigenic characterization of classical swine fever virus YC11WB isolates from wild boar
- DNA Methylation in Brain and Liver Tissues of Mice Infected with Scrapie Agent
- Classical natural ovine scrapie prions detected in practical volumes of blood by lamb and transgenic mouse bioassays
- Genetic Characteristics and Relatedness of Imported Vibrio cholerae O1 Biotype El Tor in Korea
- Re-transmissibility of mouse-adapted ME7 scrapie strain to ovine PrP transgenic mice