Anat Cell Biol.
2011 Jun;44(2):98-105. 10.5115/acb.2011.44.2.98.
Gene expression profiling of mouse aborted uterus induced by lipopolysac charide
- Affiliations
-
- 1Department of Anatomy, Research Institution of Medical Science, School of Medicine, Chonnam National University, Gwangju, Korea. atlas@jnu.ac.kr
- KMID: 1447419
- DOI: http://doi.org/10.5115/acb.2011.44.2.98
Abstract
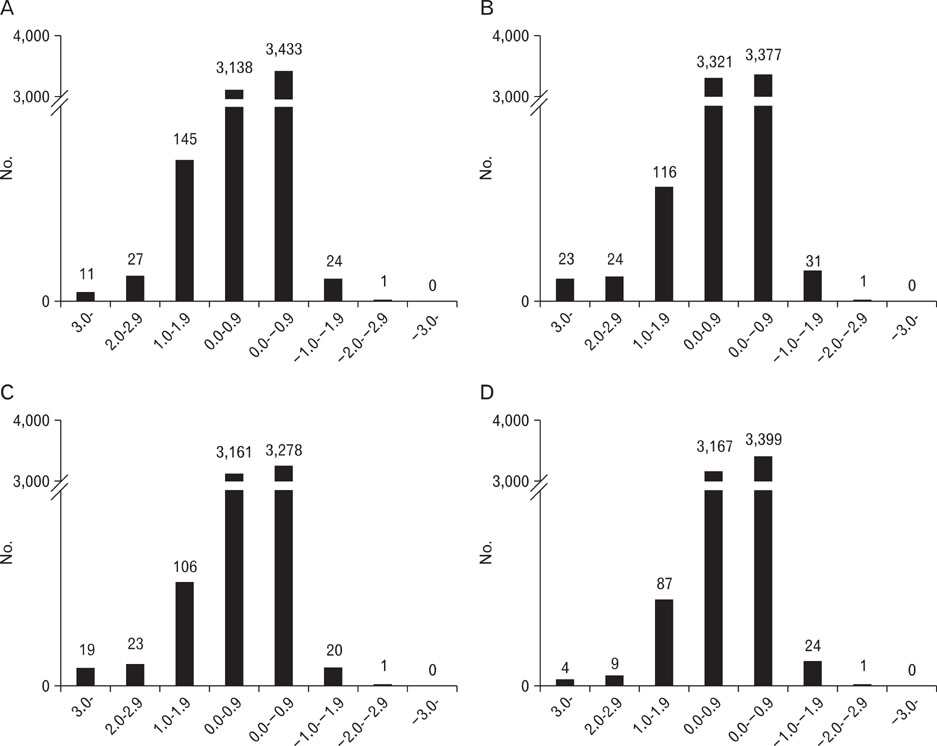
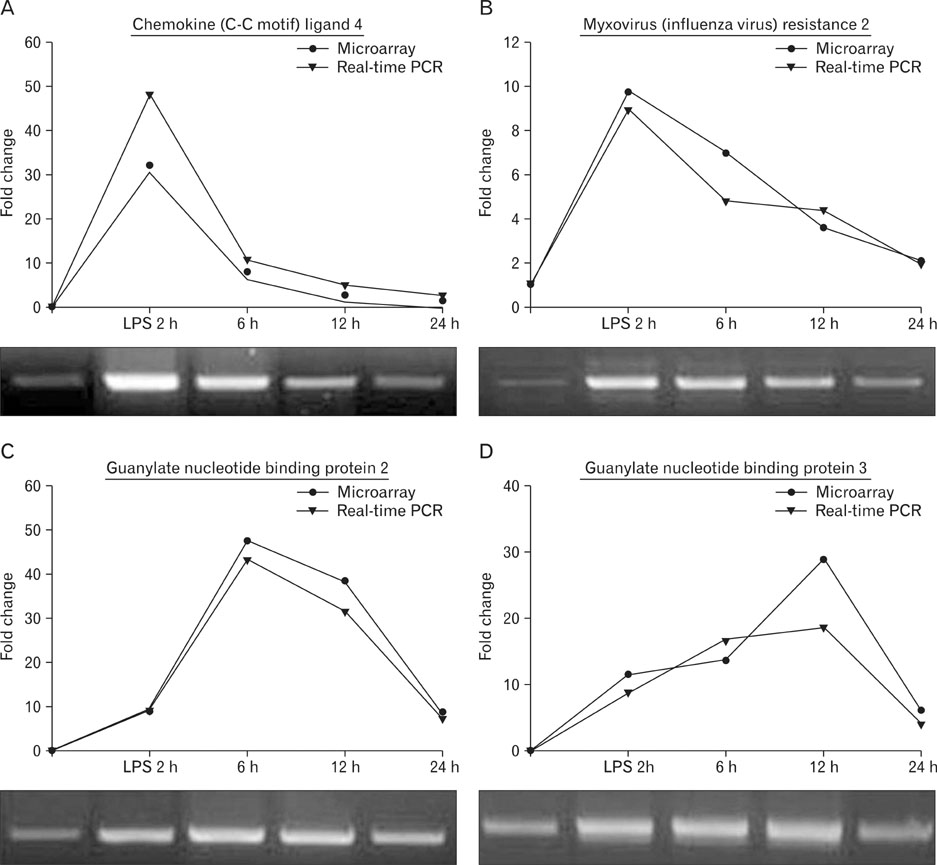
- To identify genes that participate in the abortion process, normal pregnant uteri were compared to lipopolysaccharide (LPS)-induced abortion uteri. At day 6 of pregnancy, mice were treated with LPS at various time points to induce an abortion. Total RNAs were applied to a cDNA microarray to analyze genes with altered expression. At the early stage (2 hours) of LPS-induced abortion, upregulated genes were mainly composed of immune responsive genes, including Ccl4, Ccl2, Cxcl13, Gbp3, Gbp2, Mx2, H2-Eb1, Irf1 and Ifi203. Genes related to toll-like receptor signaling were also overexpressed. At late stages of abortion (12-24 hours), many genes were suppressed rather than activated, and these were mainly related to the extracellular matrix, cytoskeleton, and anti-apoptosis. Altered expression of several selected genes was confirmed by real time reverse transcription-polymerase chain reaction. The results demonstrated that many known genes were altered in the LPS-treated pregnant uterus, implying that the molecular mechanisms of the genes involved in LPS-induced abortion are complicated. Further analysis of this expression profile will help our understanding of the pathophysiological basis for abortion.
Keyword
MeSH Terms
Figure
Reference
-
1. Blumenfeld Z, Brenner B. Thrombophilia-associated pregnancy wastage. Fertil Steril. 1999. 72:765–774.2. Li TC, Makris M, Tomsu M, Tuckerman E, Laird S. Recurrent miscarriage: aetiology, management and prognosis. Hum Reprod Update. 2002. 8:463–481.3. Laird SM, Tuckerman EM, Cork BA, Linjawi S, Blakemore AI, Li TC. A review of immune cells and molecules in women with recurrent miscarriage. Hum Reprod Update. 2003. 9:163–174.4. Zhang MQ. Large-scale gene expression data analysis: a new challenge to computational biologists. Genome Res. 1999. 9:681–688.5. Zhang X, Chen Z, Huang H, Gordon JR, Xiang J. DNA microarray analysis of the gene expression profiles of naïve versus activated tumor-specific T cells. Life Sci. 2002. 71:3005–3017.6. Lee KH, Chang MY, Ahn JI, Yu DH, Jung SS, Choi JH, Noh YH, Lee YS, Ahn MJ. Differential gene expression in retinoic acid-induced differentiation of acute promyelocytic leukemia cells, NB4 and HL-60 cells. Biochem Biophys Res Commun. 2002. 296:1125–1133.7. Deb K, Chaturvedi MM, Jaiswal YK. A 'minimum dose' of lipopolysaccharide required for implantation failure: assessment of its effect on the maternal reproductive organs and interleukin-1alpha expression in the mouse. Reproduction. 2004. 128:87–97.8. Muzikova E, Clark DA. Is spontaneous resorption in the DBA/2-mated CBA/J mouse due to a defect in "seed" or in "soil"? Am J Reprod Immunol. 1995. 33:81–85.9. Clark DA, Banwatt D, Chaouat G. Stress-triggered abortion in mice prevented by alloimmunization. Am J Reprod Immunol. 1993. 29:141–147.10. Gallinelli A, Roncaglia R, Matteo ML, Ciaccio I, Volpe A, Facchinetti F. Immunological changes and stress are associated with different implantation rates in patients undergoing in vitro fertilization-embryo transfer. Fertil Steril. 2001. 76:85–91.11. Reese J, Das SK, Paria BC, Lim H, Song H, Matsumoto H, Knudtson KL, DuBois RN, Dey SK. Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J Biol Chem. 2001. 276:44137–44145.12. Yoshioka K, Matsuda F, Takakura K, Noda Y, Imakawa K, Sakai S. Determination of genes involved in the process of implantation: application of GeneChip to scan 6500 genes. Biochem Biophys Res Commun. 2000. 272:531–538.13. Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991. 349:117–127.14. Kim S, Choi Y, Bazer FW, Spencer TE. Identification of genes in the ovine endometrium regulated by interferon tau independent of signal transducer and activator of transcription 1. Endocrinology. 2003. 144:5203–5214.15. Hsu SY, Hsueh AJ. Discovering new hormones, receptors, and signaling mediators in the genomic era. Mol Endocrinol. 2000. 14:594–604.16. Popovici RM, Kao LC, Giudice LC. Discovery of new inducible genes in in vitro decidualized human endometrial stromal cells using microarray technology. Endocrinology. 2000. 141:3510–3513.17. Riesewijk A, Martín J, van Os R, Horcajadas JA, Polman J, Pellicer A, Mosselman S, Simón C. Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Mol Hum Reprod. 2003. 9:253–264.18. Giudice LC. Elucidating endometrial function in the postgenomic era. Hum Reprod Update. 2003. 9:223–235.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gene expression profiling of mouse cavernous endothelial cells for diagnostic targets in diabetes-induced erectile dysfunction
- Gene Expression Profiling of 6-MP (6-mercaptopurine) in Liver
- Insights into the signal transduction pathways of mouse lung type II cells revealed by transcription factor profiling in the transcriptome
- Effect of Steroid Hormones on the Expression of c-Fos, CREB, ATF, and HSP70 in Rat Uterus
- Letter to the editor: Gene expression profiling of mouse cavernous endothelial cells for diagnostic targets in diabetes-induced erectile dysfunction