Korean J Ophthalmol.
2007 Jun;21(2):111-119. 10.3341/kjo.2007.21.2.111.
Microarray for Genes Associated with Signal Transduction in Diabetic OLETF Keratocytes
- Affiliations
-
- 1Department of Ophthalmology, College of Medicine, Pusan National University, Pusan, Korea. jongsool@pusan.ac.kr
- 2Siloam Eye Clinic, Pusan, Korea.
- KMID: 1101914
- DOI: http://doi.org/10.3341/kjo.2007.21.2.111
Abstract
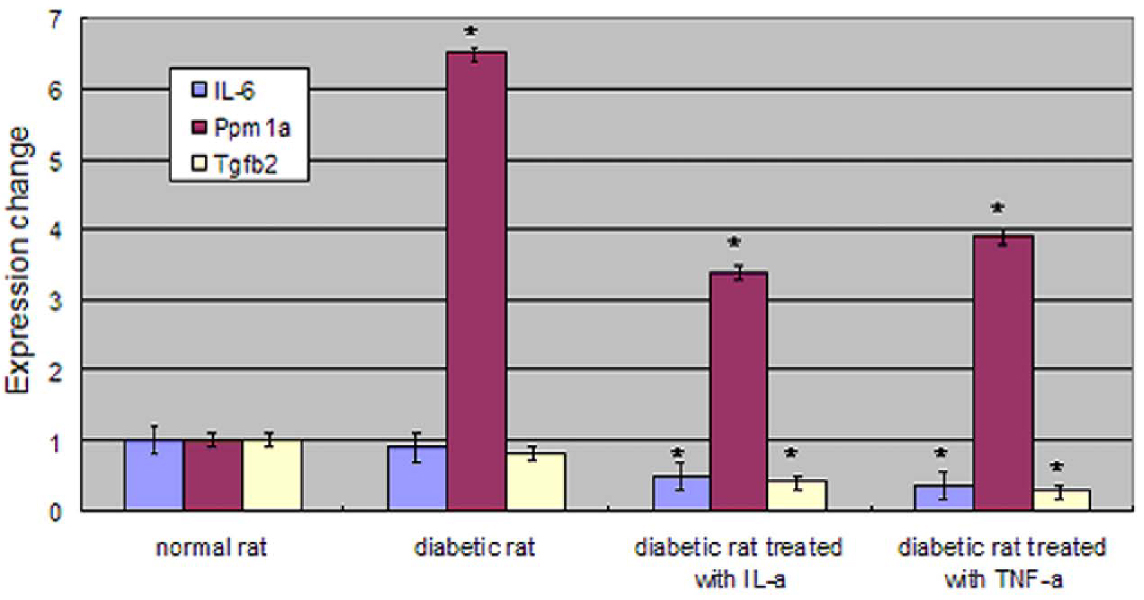
- PURPOSE: The purpose of this study was to identify differences in signal transduction gene expression between normal and diabetic keratocytes stimulated with interleukin-1alpha (IL-1alpha) and tumor necrosis factor-alpha (TNF-alpha). METHODS: Normal and diabetic keratocytes were primarily cultured and treated with 20 ng/ml IL-1alpha and TNF-alpha for 6 h. cDNA was hybridized to an oligonucleotide microarray. Genes identified by the microarray were further evaluated by real-time PCR. RESULTS: Diabetic keratocytes over-expressed components of the MAPK and Notch pathways, and under-expressed components of the insulin, calcium, and TGF-beta pathways. Cytokine treated diabetic keratocytes differentially expressed components of the TGF-beta and MAPK pathways. After IL-1alpha and TNF-alpha treatment, nine genes were under-expressed, falling in the insulin, TGF-beta, and Toll-like receptor pathways. Real-time PCR showed a significant decrease in the IL-6 and TGF-beta2 genes and a significant increase in the Ppm1a gene. CONCLUSIONS: There were some differences in gene expression between normal and diabetic keratocytes related to signal transduction pathways, such as the insulin, MAPK, calcium, and TGF-beta pathways. In addition, IL-1alpha and TNF-alpha stimulating the insulin, TGF-beta, and Toll-like receptor signaling pathways may have different effects in diabetic keratocytes.
Keyword
MeSH Terms
-
Animals
Apoptosis
Cells, Cultured
Cornea/drug effects/*metabolism/pathology
DNA/*genetics
Diabetes Mellitus, Experimental/*genetics/pathology
Gene Expression Profiling
Insulin/genetics
Interleukin-1alpha/pharmacology
Mitogen-Activated Protein Kinase Kinases/genetics
Nuclear Proteins/genetics
Oligonucleotide Array Sequence Analysis/*methods
Phosphoric Monoester Hydrolases/genetics
Polymerase Chain Reaction
Prolactin/genetics
Rats
Rats, Long-Evans
Receptors, Notch/genetics
Signal Transduction/drug effects/*genetics
Transforming Growth Factor beta/genetics
Tumor Necrosis Factor-alpha/pharmacology
Ubiquitin-Protein Ligases/genetics
Figure
Reference
-
1. Aiello LP, Garaner TW, King GL, et al. Diabetic retinopathy. Diabetes Care. 1998. 21:143–156.2. Herse PR. A review of manifestations of diabetes mellitus in the anterior eye and cornea. Am J Optom Physiol Opt. 1988. 65:224–230.3. Inoue K, Kato S, Ohara C, et al. Ocular and systemic factors relevant to diabetic keratoepitheliopathy. Cornea. 2001. 20:798–801.4. Foulks GN, Thoft RA, Perry HD, Tolentino FI. Factors related to corneal epithelial complications after closed vitrectomy in diabetes. Arch Ophthalmol. 1979. 97:1076–1078.5. Dogru M, Katakami C, Inoue M. Tear function and ocular surface changes in noninsulin-dependent diabetes mellitus. Ophthalmology. 2001. 108:586–592.6. Shultz RO, Van Hom DL, Peters MA, et al. Diabetic keratopathy. Trans Am Ophthalmol Soc. 1981. 79:180–199.7. Msuda A, Suzuki Y, Honda G, et al. Large-scale identification and characterization of human genes that activate NF-κ B and Mapk signaling pathways. Oncogene. 2003. 22:3307–3318.8. Cavllerano J. Ocular manifestation of diabetes mellitus. Optom Clin. 1992. 2:93–116.9. Cubitt CL, Lausch RN, Oakes JE. Differences in interleukin 6 gene expression between cultured human corneal epithelial cells and keratocytes. Invest Ophthalmol Vis Sci. 1995. 36:330–336.10. Singh SR, Chen X, Hou SX. JAK/STAT signaling regulates tissue outgrowth and male germline stem cell fate in Drosophilia. Cell Res. 2005. 15:1–5.11. Khodadoust AA, Silverstein AM, Kenyon K, et al. Adhesion of regeneration corneal epithelium. Invest Ophthalmol Vis Sci. 1981. 21:317–321.12. Wilson SE, Walker JW, Chwang EL, He YG. Hepatocyte growth factor (HGF), keratinocyate growth factor (KGF), their receptors, FGF receptor-2, and the cells of the cornea. Invest Ophthalmol Vis Sci. 1993. 34:2544–2561.13. Wilson SE. Role of apoptosis in wound healing in the cornea. Cornea. 2000. 19:S7–S12.14. Strissel KJ, Rinehart WB, Fini ME. A corneal epithelial inhibitor of stromal cell collagenase synthesis identified as TGF-β2. Invest Ophthalmol Vis Sci. 1995. 36:151–162.15. Shanely LJ, McCaig CD, Forrester JV, Zhao M. Insulin, not leptin, promotes in vitro cell migration to heal monolayer wounds in human corneal epithelium. Invest Ophthalmol Vis Sci. 2004. 45:1088–1094.16. Wang Y, Zhang J, Yi X, Yu FX. Activation of ERK1/2 MAP kinase pathway induces tight junction disruption in human corneal epithelial cells. Exp Eye Res. 2004. 78:125–136.17. Luo L, Li D, Doshi A, et al. Experimental dry eye stimulates production of inflammatory cytokines and MMP-9 and activates MAPK signaling pathways on the ocular surface. Invest Ophthalmol Vis Sci. 2004. 45:4293–4301.18. Klepeis VE, Cornell-Bell A, Trinkaus-Randall V. Growth factors but not gap junctions play a role in injury-induced Ca2+ waves in epithelial cells. J Cell Sci. 2001. 114:4185–4195.19. Berridge MJ, Dupont G. Spatial and temporal signaling by calcium. Curr Opin Cell Biol. 1994. 6:267–274.20. Zieske JD, Hutcheon AE, Guo X, et al. TGF-beta receptor types I and II are differentially expressed during corneal epithelial wound repair. Invest Ophthalmol Vis Sci. 2001. 42:1465–1471.21. You L, Kruse FE. Differential effect of activin A and BMP-7 on myofibroblast differentiation and the role of the Smad signaling pathway. Invest Ophthalmol Vis Sci. 2002. 43:72–81.22. Reynolds-Kenneally J, Moldzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005. 285:38–48.23. Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak-STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004. 39:141–153.24. Zhong Z, Wen Z, Darnell JE. STAT3: a STAT family member activated by tyrosine and interleukin-6. Science. 1994. 264:95–98.25. Rodringuez J, Esteve P, Weini C, et al. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005. 8:1301–1309.26. Li CH, Amar S. Role of Secreted Frizzled-related Protein 1 (SFRP1) in Wound Healing. J Dent Res. 2006. 85:374–378.27. Kida T, Ikeda T, Nishimura M, et al. Renin-angiotensin system in proliferative diabetic retinopathy and its gene expression in cultured human muller cells. Jpn J Ophthalmol. 2003. 47:36–41.28. Mizoue S, Iwai M, ide A, et al. Role of angiotensin II receptor subtypes in conjunctival wound healing. Curr Eye Res. 2006. 31:129–136.29. Zhang J, Xu K, Ambati B, Yu FX. Toll-like receptor 5-mediated corneal epithelial inflammatory responses to pseudomonas aeruginosa flagellin. Invest Ophthalmol Vis Sci. 2003. 44:4247–4254.30. Johnson AC, Heinzel FP, Diaconu E, et al. Activation of toll-like receptor (TLR)2, TLR4, and TLR9 in the mammalian cornea induces MyD88-dependent corneal inflammation. Invest Ophthalmol Vis Sci. 2005. 46:589–595.31. Chen SH, Oakes JE, lausch RN. Synergistic anti-herpes effect of TNF-alpha and IFN-gamma in human corneal epithelial cells compared with that in corneal fibroblasts. Antiviral Res. 1994. 25:201–213.32. Ekstrand AJ, Cao R, Bjorndahl M, et al. Deletion of neuropeptide Y (NPY) 2 receptor in mice results in blockage of NPY-induced angiogenesis and delayed wound healing. Proc Natl Acad Sci U S A. 2003. 100:6033–6038.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gene Microarray Related with Apoptosis in Diabetic OLETF Keratocytes
- Microarray Analysis for Genes Associated with Angiogenesis in Diabetic OLETF Keratocytes
- Profile of Gene Expression Changes During Doxorubicin Induced Apoptosis of Saos-2
- Bone/Vascular Calcification: Signal Transduction Pathway and Calcification Related Genes
- Analysis of Gene Expression in the Human Chorion of Preterm Labor Using cDNA Microarray