Korean J Lab Med.
2011 Apr;31(2):61-71. 10.3343/kjlm.2011.31.2.61.
The Path to Clinical Proteomics Research: Integration of Proteomics, Genomics, Clinical Laboratory and Regulatory Science
- Affiliations
-
- 1Office of Cancer Clinical Proteomics Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA. rodriguezh@mail.nih.gov
- KMID: 1094342
- DOI: http://doi.org/10.3343/kjlm.2011.31.2.61
Abstract
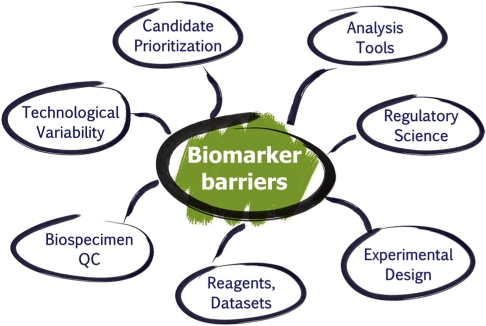
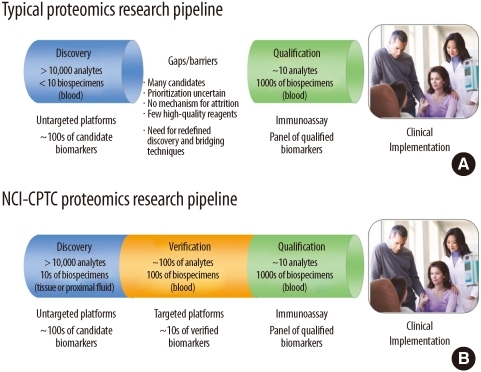
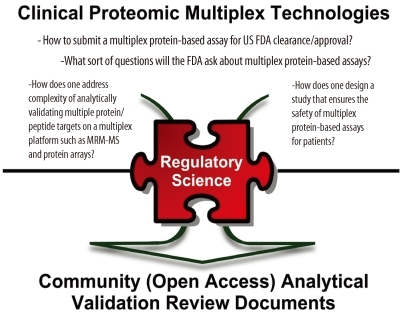
- Better biomarkers are urgently needed to cancer detection, diagnosis, and prognosis. While the genomics community is making significant advances in understanding the molecular basis of disease, proteomics will delineate the functional units of a cell, proteins and their intricate interaction network and signaling pathways for the underlying disease. Great progress has been made to characterize thousands of proteins qualitatively and quantitatively in complex biological systems by utilizing multi-dimensional sample fractionation strategies, mass spectrometry and protein microarrays. Comparative/quantitative analysis of high-quality clinical biospecimen (e.g., tissue and biofluids) of human cancer proteome landscape has the potential to reveal protein/peptide biomarkers responsible for this disease by means of their altered levels of expression, post-translational modifications as well as different forms of protein variants. Despite technological advances in proteomics, major hurdles still exist in every step of the biomarker development pipeline. The National Cancer Institute's Clinical Proteomic Technologies for Cancer initiative (NCI-CPTC) has taken a critical step to close the gap between biomarker discovery and qualification by introducing a pre-clinical "verification" stage in the pipeline, partnering with clinical laboratory organizations to develop and implement common standards, and developing regulatory science documents with the US Food and Drug Administration to educate the proteomics community on analytical evaluation requirements for multiplex assays in order to ensure the safety and effectiveness of these tests for their intended use.
MeSH Terms
Figure
Cited by 1 articles
-
Proteomic Profiling of Serum from Patients with Tuberculosis
Sang Hoon Song, Minje Han, Yang Seon Choi, Ki Soon Dan, Man Gil Yang, Junghan Song, Sung Sup Park, Jae Ho Lee
Ann Lab Med. 2014;34(5):345-353. doi: 10.3343/alm.2014.34.5.345.
Reference
-
1. Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, et al. The case for early detection. Nat Rev Cancer. 2003; 3:243–252. PMID: 12671663.
Article2. Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005; 563:23–60. PMID: 15611012.
Article3. Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006; 24:971–983. PMID: 16900146.
Article4. García-Foncillas J, Bandrés E, Zárate R, Remírez N. Proteomic analysis in cancer research: potential application in clinical use. Clin Transl Oncol. 2006; 8:250–261. PMID: 16648100.
Article5. Bouchal P, Roumeliotis T, Hrstka R, Nenutil R, Vojtesek B, Garbis SD. Biomarker discovery in low-grade breast cancer using isobaric stable isotope tags and two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2DLC-MS/MS) based quantitative proteomic analysis. J Proteome Res. 2009; 8:362–373. PMID: 19053527.
Article6. Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004; 76:6085–6096. PMID: 15481957.
Article7. Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 2010; 7:383–385. PMID: 20364148.
Article8. Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006; 5:573–588. PMID: 16332733.
Article9. Wang H, Wong CH, Chin A, Kennedy J, Zhang Q, Hanash S. Quantitative serum proteomics using dual stable isotope coding and nano LC-MS/MSMS. J Proteome Res. 2009; 8:5412–5422. PMID: 19817497.
Article10. Lee J, Soper SA, Murray KK. Microfluidic chips for mass spectrometry-based proteomics. J Mass Spectrom. 2009; 44:579–593. PMID: 19373851.
Article11. Pierobon M, Calvert V, Belluco C, Garaci E, Deng J, Lise M, et al. Multiplexed cell signaling analysis of metastatic and nonmetastatic colorectal cancer reveals COX2-EGFR signaling activation as a potential prognostic pathway biomarker. Clin Colorectal Cancer. 2009; 8:110–117.
Article12. Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, et al. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008; 5:535–538. PMID: 18469824.
Article13. Beirne P, Pantelidis P, Charles P, Wells AU, Abraham DJ, Denton CP, et al. Multiplex immune serum biomarker profiling in sarcoidosis and systemic sclerosis. Eur Respir J. 2009; 34:1376–1382. PMID: 19541722.
Article14. Kelleher MT, Fruhwirth G, Patel G, Ofo E, Festy F, Barber PR, et al. The potential of optical proteomic technologies to individualize prognosis and guide rational treatment for cancer patients. Target Oncol. 2009; 4:235–252. PMID: 19756916.
Article15. Wang P, Whiteaker JR, Paulovich AG. The evolving role of mass spectrometry in cancer biomarker discovery. Cancer Biol Ther. 2009; 8:1083–1094. PMID: 19502776.
Article16. Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng LC, et al. Head-to-head comparison of serum fractionation techniques. J Proteome Res. 2007; 6:828–836. PMID: 17269739.
Article17. Ernoult E, Bourreau A, Gamelin E, Guette C. A proteomic approach for plasma biomarker discovery with iTRAQ labeling and OFFGEL fractionation. J Biomed Biotechnol. 2010; 2010:927917. PMID: 19888438.18. Nirmalan NJ, Hughes C, Peng J, McKenna T, Langridge J, Cairns DA, et al. Initial development and validation of a novel extraction method for quantitative mining of the formalin-fixed, paraffin-embedded tissue proteome for biomarker investigations. J Proteome Res. 2010; 10:896–906. PMID: 21117664.
Article19. Krishhan VV, Khan IH, Luciw PA. Multiplexed microbead immunoassays by flow cytometry for molecular profiling: basic concepts and proteomics applications. Crit Rev Biotechnol. 2009; 29:29–43. PMID: 19514901.
Article20. Cha S, Imielinski MB, Rejtar T, Richardson EA, Thakur D, Sgroi DC, et al. In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: annotation by protein set enrichment analysis and gene ontology. Mol Cell Proteomics. 2010; 9:2529–2544. PMID: 20739354.
Article21. Anderson KS, Sibani S, Wallstrom G, Qiu J, Mendoza EA, Raphael J, et al. Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J Proteome Res. 2011; 10:85–96. PMID: 20977275.
Article22. Bateman NW, Sun M, Hood BL, Flint MS, Conrads TP. Defining central themes in breast cancer biology by differential proteomics: conserved regulation of cell spreading and focal adhesion kinase. J Proteome Res. 2010; 9:5311–5324. PMID: 20681588.
Article23. Kristiansen TZ, Harsha HC, Grønborg M, Maitra A, Pandey A. Differential membrane proteomics using 18O-labeling to identify biomarkers for cholangiocarcinoma. J Proteome Res. 2008; 7:4670–4677. PMID: 18839982.24. An HJ, Lebrilla CB. A glycomics approach to the discovery of potential cancer biomarkers. Methods Mol Biol. 2010; 600:199–213. PMID: 19882130.
Article25. Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010; 11:427–439. PMID: 20461098.
Article26. Madian AG, Regnier FE. Profiling carbonylated proteins in human plasma. J Proteome Res. 2010; 9:1330–1343. PMID: 20121119.
Article27. Iwabata H, Yoshida M, Komatsu Y. Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics. 2005; 5:4653–4664. PMID: 16247734.
Article28. Ceroni A, Sibani S, Baiker A, Pothineni VR, Bailer SM, LaBaer J, et al. Systematic analysis of the IgG antibody immune response against varicella zoster virus (VZV) using a self-assembled protein microarray. Mol Biosyst. 2010; 6:1604–1610. PMID: 20514382.
Article29. Wong J, Sibani S, Lokko NN, LaBaer J, Anderson KS. Rapid detection of antibodies in sera using multiplexed self-assembling bead arrays. J Immunol Methods. 2009; 350:171–182. PMID: 19732778.
Article30. Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010; 56:177–185. PMID: 19884488.
Article31. Osterfeld SJ, Yu H, Gaster RS, Caramuta S, Xu L, Han SJ, et al. Multiplex protein assays based on real-time magnetic nanotag sensing. Proc Natl Acad Sci USA. 2008; 105:20637–20640. PMID: 19074273.
Article32. Paulovich AG, Billheimer D, Ham AJ, Vega-Montoto L, Rudnick PA, Tabb DL, et al. Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS platform performance. Mol Cell Proteomics. 2010; 9:242–254. PMID: 19858499.
Article33. Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blonder N, et al. Performance metrics for liquid chromatography-tandem mass spectrometry systems in proteomics analyses. Mol Cell Proteomics. 2010; 9:225–241. PMID: 19837981.
Article34. Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009; 27:633–641. PMID: 19561596.
Article35. Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010; 9:761–776. PMID: 19921851.
Article36. Rodriguez H, Snyder M, Uhlén M, Andrews P, Beavis R, Borchers C, et al. Recommendations from the 2008 international summit on proteomics data release and sharing policy: the amsterdam principles. J Proteome Res. 2009; 8:3689–3692. PMID: 19344107.
Article37. Gloriam DE, Orchard S, Bertinetti D, Björling E, Bongcam-Rudloff E, Borrebaeck CA, et al. A community standard format for the representation of protein affinity reagents. Mol Cell Proteomics. 2010; 9:1–10. PMID: 19674966.
Article38. Xu X, Zhang J, Zhang L, Liu W, Weisel CP. Selective detection of monohydroxy metabolites of polycyclic aromatic hydrocarbons in urine using liquid chromatography/triple quadrupole tandem mass spectrometry. Rapid Commun Mass Spectrom. 2004; 18:2299–2308. PMID: 15384151.
Article39. Xu X, Roman JM, Issaq HJ, Keefer LK, Veenstra TD, Ziegler RG. Quantitative measurement of endogenous estrogens and estrogen metabolites in human serum by liquid chromatography-tandem mass spectrometry. Anal Chem. 2007; 79:7813–7821. PMID: 17848096.
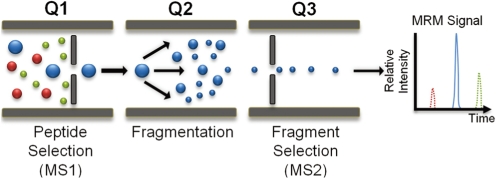
Article40. James A, Jorgensen C. Basic design of MRM assays for peptide quantification. Methods Mol Biol. 2010; 658:167–185. PMID: 20839104.
Article41. Kiyonami R, Schoen A, Prakash A, Peterman S, Zabrouskov V, Picotti P, et al. Increased selectivity, analytical precision, and throughput in targeted proteomics. Mol Cell Proteomics. 2011; 10:M110.002931. PMID: 20664071.
Article42. Gerszten RE, Carr SA, Sabatine M. Integration of proteomic-based tools for improved biomarkers of myocardial injury. Clin Chem. 2010; 56:194–201. PMID: 20022985.
Article43. Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004; 4:1175–1186. PMID: 15048997.44. Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, et al. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem. 2009; 55:1108–1117. PMID: 19372185.
Article45. Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA). J Proteome Res. 2004; 3:235–244. PMID: 15113099.
Article46. Ahn YH, Lee JY, Lee JY, Kim YS, Ko JH, Yoo JS. Quantitative analysis of an aberrant glycoform of TIMP1 from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J Proteome Res. 2009; 8:4216–4224. PMID: 19645485.
Article47. Reid JD, Holmes DT, Mason DR, Shah B, Borchers CH. Towards the development of an immuno MALDI (iMALDI) mass spectrometry assay for the diagnosis of hypertension. J Am Soc Mass Spectrom. 2010; 21:1680–1686. PMID: 20199871.
Article48. Jiang J, Parker CE, Fuller JR, Kawula TH, Borchers CH. An immunoaffinity tandem mass spectrometry (iMALDI) assay for detection of Francisella tularensis. Anal Chim Acta. 2007; 605:70–79. PMID: 18022413.
Article49. Mansfield E, O'Leary TJ, Gutman SI. Food and drug administration regulation of in vitro diagnostic devices. J Mol Diagn. 2005; 7:2–7. PMID: 15681468.
Article50. FDA/CDRH Public Meeting: oversight of Laboratory Developed Tests (LDTs), July 19-20, 2010. U.S. Food and Drug Administration (FDA). Updated on Sep 2010. http://www.fda.gov/MedicalDevices/NewsEvents/WorkshopsConferences/ucm212830.htm.51. Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010; 28:827–838. PMID: 20676074.
Article52. Draft guidance for industry, clinical laboratories, and FDA staff: in vitro diagnostic multivariate index assays. Center for Devices and Radiological Health (CDRH). Updated on Jul 2007. http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm071455.pdf.53. 510(k) Substantial equivalence determination decision summary. U.S. Food and drug administration. Updated on Mar 2011. http://www.accessdata.fda.gov/cdrh_docs/reviews/K081754.pdf.54. Rodriguez H, Tezak Z, Mesri M, Carr SA, Liebler DC, Fisher SJ, et al. Analytical validation of protein-based multiplex assays: a workshop report by the NCI-FDA interagency oncology task force on molecular diagnostics. Clin Chem. 2010; 56:237–243. PMID: 20007859.
Article55. Regnier FE, Skates SJ, Mesri M, Rodriguez H, Tezak Z, Kondratovich MV, et al. Protein-based multiplex assays: mock presubmissions to the US food and drug administration. Clin Chem. 2010; 56:165–171. PMID: 20007858.
Article56. Pacini F. Follow-up of differentiated thyroid cancer. Eur J Nucl Med Mol Imaging. 2002; 29(S2):S492–S496. PMID: 12192551.
Article57. Saghari M, Gholamrezanezhad A, Mirpour S, Eftekhari M, Takavar A, Fard-Esfahani A, et al. Efficacy of radioiodine therapy in the treatment of elevated serum thyroglobulin in patients with differentiated thyroid carcinoma and negative whole-body iodine scan. Nucl Med Commun. 2006; 27:567–572. PMID: 16794517.
Article58. Spencer CA. Recoveries cannot be used to authenticate thyroglobulin (Tg) measurements when sera contain Tg autoantibodies. Clin Chem. 1996; 42:661–663. PMID: 8653888.
Article59. Spencer CA. Challenges of serum thyroglobulin (Tg) measurement in the presence of Tg autoantibodies. J Clin Endocrinol Metab. 2004; 89:3702–3704. PMID: 15292292.
Article60. Preissner CM, O'Kane DJ, Singh RJ, Morris JC, Grebe SK. Phantoms in the assay tube: heterophile antibody interferences in serum thyroglobulin assays. J Clin Endocrinol Metab. 2003; 88:3069–3074. PMID: 12843145.
Article61. Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008; 54:1796–1804. PMID: 18801935.
Article62. Hoofnagle AN. Peptide lost and found: internal standards and the mass spectrometric quantification of peptides. Clin Chem. 2010; 56:1515–1517. PMID: 20739635.
Article63. Ciccimaro E, Hanks SK, Yu KH, Blair IA. Absolute quantification of phosphorylation on the kinase activation loop of cellular focal adhesion kinase by stable isotope dilution liquid chromatography/mass spectrometry. Anal Chem. 2009; 81:3304–3313. PMID: 19354260.
Article64. CFR-Code federal regulations title 21. U.S. Food and Drug Administration (FDA). Updated on Apr 2010. http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=862.2860.65. Clinical and Laboratory Standards Institute (CLSI). Updated on Feb 2011. http://www.clsi.org.66. Spitz MR, Bondy ML. The evolving discipline of molecular epidemiology of cancer. Carcinogenesis. 2010; 31:127–134. PMID: 20022891.
Article67. Rosa DD, Ismael G, Lago LD, Awada A. Molecular-targeted therapies: lessons from years of clinical development. Cancer Treat Rev. 2008; 34:61–80. PMID: 17826917.
Article68. Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009; 302:261–275. PMID: 19602686.
Article69. International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010; 464:993–998. PMID: 20393554.70. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–1068. PMID: 18772890.