Spectra of Chromosomal Aberrations in 325 Leukemia Patients and Implications for the Development of New Molecular Detection Systems
- Affiliations
-
- 1Department of Laboratory Medicine, Chonnam National University Hwasun Hospital, Hwasun, Korea. mgshin@chonnam.ac.kr
- 2Genome Research Center for Hematopoietic Diseases, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- 3Brain Korea 21 Project, Center for Biomedical Human Resources at Chonnam National University, Gwangju, Korea.
- 4Environmental Health Center for Childhood Leukemia and Cancer, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- KMID: 1094259
- DOI: http://doi.org/10.3346/jkms.2011.26.7.886
Abstract
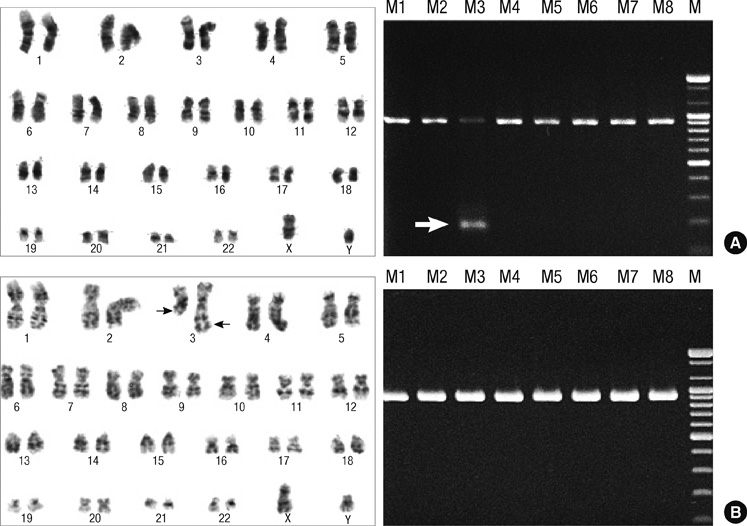
- This study investigated the spectrum of chromosomal abnormalities in 325 leukemia patients and developed optimal profiles of leukemic fusion genes for multiplex RT-PCR. We prospectively analyzed blood and bone marrow specimens of patients with acute leukemia. Twenty types of chromosomal abnormalities were detected in 42% from all patients by commercially available multiplex RT-PCR for detecting 28 fusion genes and in 35% by cytogenetic analysis including FISH analysis. The most common cytogenetic aberrations in acute myeloid leukemia patients was PML/PARA, followed by AML1/MGT8 and MLL1, and in acute lymphoid leukemia patients was BCR/ABL, followed by TEL/AML1 and MLL1 gene rearrangement. Among the negative results for multiplex RT-PCR, clinically significant t(3;3)(q21;q26.2), t(8;14)(q24;q32) and i(17)(q10) were detected by conventional cytogenetics. The spectrum and frequency of chromosomal abnormalities in our leukemia patients are differed from previous studies, and may offer optimal profiles of leukemic fusion genes for the development of new molecular detection systems.
MeSH Terms
-
Adaptor Proteins, Signal Transducing/genetics
Adult
Aged
Aged, 80 and over
*Chromosome Aberrations
Chromosomes, Human, Pair 14
Chromosomes, Human, Pair 17
Chromosomes, Human, Pair 3
Chromosomes, Human, Pair 8
Core Binding Factor Alpha 2 Subunit/genetics
Female
Fusion Proteins, bcr-abl/genetics
Gene Rearrangement
Humans
In Situ Hybridization, Fluorescence
Karyotyping
Leukemia/diagnosis/*genetics
Leukemia, Myeloid, Acute/diagnosis/genetics
Male
Middle Aged
Myeloid-Lymphoid Leukemia Protein/genetics
Oncogene Proteins, Fusion/genetics
Precursor Cell Lymphoblastic Leukemia-Lymphoma/diagnosis/genetics
Reverse Transcriptase Polymerase Chain Reaction
Figure
Cited by 4 articles
-
Changing the frequency and spectra of chromosomal aberrations in Korean patients with acute leukemia in a tertiary care hospital
Je-Hyun Park, Min-Gu Kang, Hye-Ran Kim, Young-Eun Lee, Jun Hyung Lee, Hyun-Jung Choi, Jong-Hee Shin, Myung-Geun Shin
Blood Res. 2020;55(4):225-245. doi: 10.5045/br.2020.2020255.Diagnostic Usefulness of Genomic Breakpoint Analysis of Various Gene Rearrangements in Acute Leukemias: A Perspective of Long Distance- or Long Distance Inverse-PCR-based Approaches
John Jeongseok Yang, Rolf Marschalek, Claus Meyer, Tae Sung Park
Ann Lab Med. 2012;32(4):316-318. doi: 10.3343/alm.2012.32.4.316.Alteration of the
SETBP1 Gene and Splicing Pathway GenesSF3B1 ,U2AF1 , andSRSF2 in Childhood Acute Myeloid Leukemia
Hyun-Woo Choi, Hye-Ran Kim, Hee-Jo Baek, Hoon Kook, Duck Cho, Jong-Hee Shin, Soon-Pal Suh, Dong-Wook Ryang, Myung-Geun Shin
Ann Lab Med. 2015;35(1):118-122. doi: 10.3343/alm.2015.35.1.118.Identification of Mixed Lineage Leukemia Gene (
MLL )/MLLT10 Fusion Transcripts by Reverse Transcription-PCR and Sequencing in a Case of AML With a FISH-Negative CrypticMLL Rearrangement
Kiwoong Ko, Min-Jung Kwon, Hee-Yeon Woo, Hyosoon Park, Chang-Hun Park, Seung-Tae Lee, Sun-Hee Kim
Ann Lab Med. 2015;35(4):469-471. doi: 10.3343/alm.2015.35.4.469.
Reference
-
1. Pallisgaard N, Hokland P, Riishøj DC, Pedersen B, Jørgensen P. Multiplex reverse transcription-polymerase chain reaction for simultaneous screening of 29 translocations and chromosomal aberrations in acute leukemia. Blood. 1998. 92:574–588.2. Bacher U, Kern W, Schnittger S, Hiddemann W, Schoch C, Haferlach T. Further correlations of morphology according to FAB and WHO classification to cytogenetics in de novo acute myeloid leukemia: a study on 2,235 patients. Ann Hematol. 2005. 84:785–791.3. Betts DR, Ammann RA, Hirt A, Hengartner H, Beck-Popovic M, Kuhne T, Nobile L, Caflisch U, Wacker P, Niggli FK. The prognostic significance of cytogenetic aberrations in childhood acute myeloid leukaemia. A study of the Swiss Paediatric Oncology Group (SPOG). Eur J Haematol. 2007. 78:468–476.4. Klaus M, Haferlach T, Schnittger S, Kern W, Hiddemann W, Schoch C. Cytogenetic profile in de novo acute myeloid leukemia with FAB subtypes M0, M1, and M2: a study based on 652 cases analyzed with morphology, cytogenetics, and fluorescence in situ hybridization. Cancer Genet Cytogenet. 2004. 155:47–56.5. Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997. 278:1059–1064.6. Hutchings Hoffmann M, Wirenfeldt Klausen T, Hasle H, Schmiegelow K, Brondum-Nielsen K, Johnsen HE. Multiplex reverse transcription polymerase chain reaction screening in acute myeloid leukemia detects cytogenetically unrevealed abnormalities of prognostic significance. Haematologica. 2005. 90:984–986.7. Strehl S, König M, Mann G, Haas OA. Multiplex reverse transcriptase-polymerase chain reaction screening in childhood acute myeloblastic leukemia. Blood. 2001. 97:805–808.8. Elia L, Mancini M, Moleti L, Meloni G, Buffolino S, Krampera M, De Rossi G, Foà R, Cimino G. A multiplex reverse transcriptase-polymerase chain reaction strategy for the diagnostic molecular screening of chimeric genes: a clinical evaluation on 170 patients with acute lymphoblastic leukemia. Haematologica. 2003. 88:275–279.9. Nakase K, Bradstock K, Sartor M, Gottlieb D, Byth K, Kita K, Shiku H, Kamada N. Geographic heterogeneity of cellular characteristics of acute myeloid leukemia: a comparative study of Australian and Japanese adult cases. Leukemia. 2000. 14:163–168.10. Cheng Y, Wang Y, Wang H, Chen Z, Lou J, Xu H, Qian W, Meng H, Lin M, Jin J. Cytogenetic profile of de novo acute myeloid leukemia: a study based on 1432 patients in a single institution of China. Leukemia. 2009. 23:1801–1806.11. Pui CH. Acute lymphoblastic leukemia in children. Curr Opin Oncol. 2000. 12:3–12.12. Pullarkat V, Slovak ML, Kopecky KJ, Forman SJ, Appelbaum FR. Impact of cytogenetics on the outcome of adult acute lymphoblastic leukemia: results of Southwest Oncology Group 9400 study. Blood. 2008. 111:2563–2572.13. Amare P, Gladstone B, Varghese C, Pai S, Advani S. Clinical significance of cytogenetic findings at diagnosis and in remission in childhood and adult acute lymphoblastic leukemia: experience from India. Cancer Genet Cytogenet. 1999. 110:44–53.14. Chang HH, Lu MY, Jou ST, Lin KH, Tien HF, Lin DT. Cytogenetics in childhood acute lymphoblastic leukemia in Taiwan: a single-institutional experience. Pediatr Hematol Oncol. 2006. 23:495–506.15. Pinheiro RF, Chauffaille Mde L, Silva MR. Isochromosome 17q in MDS: a marker of a distinct entity. Cancer Genet Cytogenet. 2006. 166:189–190.16. Kim MH, Hwang HY, Jeong SH, Kim YS, Eo WK, Park JS, Moon YH. Detection of p53 mutant and isochromosome 17q in myelodysplastic syndromes and leukemias. Korean J Clin Pathol. 2000. 20:349–353.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Review of Chromosomal Analyses Performed in a Single Hospital
- Assessment of Chromosomal Analyses of 1,180 Cases Suspected of Chromosomal Aberrations
- Changing the frequency and spectra of chromosomal aberrations in Korean patients with acute leukemia in a tertiary care hospital
- Additional cytogenetic aberrations in chronic myeloid leukemia: a single-center experience in the Middle East
- Chromosomal Aberrations and Molecular Cytogenetics in Cancer