J Korean Med Sci.
2011 Jul;26(7):851-858. 10.3346/jkms.2011.26.7.851.
Gene Expression Profile during Chondrogenesis in Human Bone Marrow derived Mesenchymal Stem Cells using a cDNA Microarray
- Affiliations
-
- 1Department of Internal Medicine, Rheumatism Research Institute, Seoul National University College of Medicine, Seoul, Korea. ysong@snu.ac.kr
- 2Graduate Course of Biomedical Informatics (SNUBI), Seoul National University College of Medicine, Seoul, Korea.
- KMID: 1094253
- DOI: http://doi.org/10.3346/jkms.2011.26.7.851
Abstract
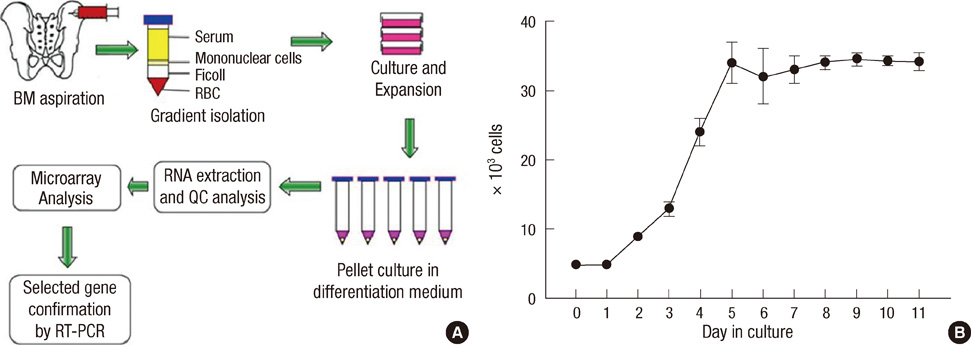
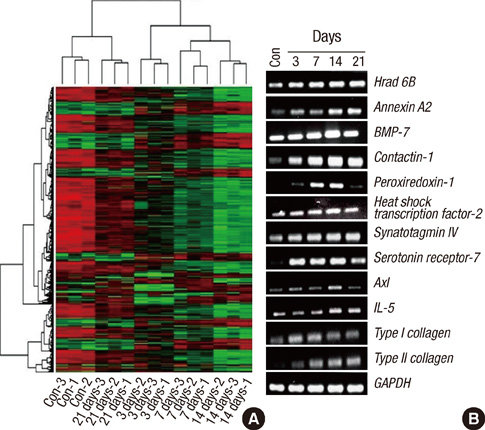
- Mesenchymal stem cells (MSCs) have the capacity to proliferate and differentiate into multiple connective tissue lineages, which include cartilage, bone, and fat. Cartilage differentiation and chondrocyte maturation are required for normal skeletal development, but the intracellular pathways regulating this process remain largely unclear. This study was designed to identify novel genes that might help clarify the molecular mechanisms of chondrogenesis. Chondrogenesis was induced by culturing human bone marrow (BM) derived MSCs in micromass pellets in the presence of defined medium for 3, 7, 14 or 21 days. Several genes regulated during chondrogenesis were then identified by reverse transcriptase-polymerase chain reaction (RT-PCR). Using an ABI microarray system, we determined the differential gene expression profiles of differentiated chondrocytes and BM-MSCs. Normalization of this data resulted in the identification of 1,486 differentially expressed genes. To verify gene expression profiles determined by microarray analysis, the expression levels of 10 genes with high fold changes were confirmed by RT-PCR. Gene expression patterns of 9 genes (Hrad6B, annexinA2, BMP-7, contactin-1, peroxiredoxin-1, heat shock transcription factor-2, synaptotagmin IV, serotonin receptor-7, Axl) in RT-PCR were similar to the microarray gene expression patterns. These findings provide novel information concerning genes involved in the chondrogenesis of human BM-MSCs.
MeSH Terms
Figure
Reference
-
1. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999. 284:143–147.2. Ho YC, Chung YC, Hwang SM, Wang KC, Hu YC. Transgene expression and differentiation of baculovirus-transduced human mesenchymal stem cells. J Gene Med. 2005. 7:860–868.3. Haynesworth SE, Baber MA, Caplan AI. Cell surface antigens on human marrow-derived mesenchymal cells are detected by monoclonal antibodies. Bone. 1992. 13:69–80.4. Izzo MW, Pucci B, Tuan RS, Hall DJ. Gene expression profiling following BMP-2 induction of mesenchymal chondrogenesis in vitro. Osteoarthritis Cartilage. 2002. 10:23–33.5. Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqudam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996. 93:6025–6030.6. Bosotti R, Locatelli G, Healy S, Scacheri E, Sartori L, Mercurio C, Calogero R, Isacchi A. Cross platform microarray analysis for robust identification of differentially expressed genes. BMC Bioinformatics. 2007. 8:Suppl 1. S5.7. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004. 5:R80.8. Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002. 32:Suppl. 496–501.9. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998. 95:14863–14868.10. Goruppi S, Chiaruttini C, Ruaro ME, Varnum B, Schneider C. Gas6 induces growth, beta-catenin stabilization, and T-cell factor transcriptional activation in contact-inhibited C57 mammary cells. Mol Cell Biol. 2001. 21:902–915.11. Budagian V, Bulanova E, Orinska Z, Thon L, Mamat U, Bellosta P, Adam D, Paus R, Bulfone-Paus S. A promiscuous liaison between IL-15 receptor and Axl receptor tyrosine kinase in cell death control. EMBO J. 2005. 24:4260–4270.12. Ahras M, Otto GP, Tooze SA. Synaptotagmin IV is necessary for the maturation of secretory granules in PC12 cells. J Cell Biol. 2006. 173:241–251.13. Sasaki H, Moriyama S, Nakashima Y, Yukiue H, Fukai I, Fuji Y. Decreased Hrad6B expression in lung cancer. Acta Oncol. 2004. 43:585–589.14. Immenschuh S, Baugmart-Vogt E, Tan M, Iwahara SI, Ramadori G, Fahimi HD. Differential cellular and subcellular localization of heme-binding protein 23/peroxiredoxin I and heme oxygenase-1 in rat liver. J Histochem Cytochem. 2003. 51:1621–1631.15. Leyens G, Donnay I, Knoops B. Cloning of bovine peroxiredoxins-gene expression in bovine tissues and amino acid sequence comparison with rat, mouse and primate peroxiredoxins. Comp Biochem Physiol B Biochem Mol Biol. 2003. 136:943–955.16. Bai X, Li G, Zhao C, Duan H, Qu F. BMP7 induces the differentiation of bone marrow-derived mesenchymal cells into chondrocytes. Med Biol Eng Comput. 2011. 49:687–692.17. Eriksson M, Jokinen E, Sistonen L, Leppä S. Heat shock factor 2 is activated during mouse heart development. Int J Dev Biol. 2000. 44:471–477.18. Kajiya H, Ito M, Ohshima H, Kenmotsu S, Ries WL, Benjamin IJ, Reddy SV. RANK ligand expression in heat shock factor-2 deficient mouse bone marrow stromal/preosteoblast cells. J Cell Biochem. 2006. 97:1362–1369.19. Dreier R, Schmid KW, Greke V, Riehemann K. Differential expression of annexins I, II and IV in human tissues: an immunohistochemical study. Histochem Cell Biol. 1998. 110:137–148.20. Diaz VM, Hurtado M, Thomson TM, Reventós J, Paciucci R. Specific interaction of tissue-type plasminogen activator (t-PA) with annexin II on the membrane of pancreatic cancer cells activates plasminogen and promotes invasion in vitro. Gut. 2004. 53:993–1000.21. Fang YT, Lin CF, Liao PC, Kuo YM, Wang S, Yeh TM, Shieh CC, Su IJ, Lei HY, Lin YS. Annexin A2 on lung epithelial cell surface is recognized by severe acute respiratory syndrome-associated coronavirus spike domain 2 antibodies. Mol Immunol. 2010. 47:1000–1009.22. Esposito I, Penzel R, Chaib-Harrireche M, Barcena U, Bergmann F, Riedl S, Kayed H, Giese N, Kleeff J, Freiss H, Schirmacher P. Tenascin C and annexin II expression in the process of pancreatic carcinogenesis. J Pathol. 2006. 208:673–685.23. Tanaka T, Akatsuka S, Ozeki M, Shirase T, Hiai H, Toyokuni S. Redox regulation of annexin 2 and its implication for oxidative stress-induced renal carcinogenesis and metastasis. Oncogene. 2004. 23:3980–3989.24. Falk J, Bonnon C, Girault JA, Faivre-Sarrailh C. F3/contactin, a neuronal cell adhesion molecule implicated in axogenesis and myelination. Biol Cell. 2002. 94:327–334.25. Eckerich C, Zapf S, Ulbricht U, Müller S, Fillbrandt R, Westphal M, Lamszus K. Contactin is expressed in human astrocytic gliomas and mediates repulsive effects. Glia. 2006. 53:1–12.26. Majumdar MK, Thiede MA, Mosca JD, Moorman M, Gerson SL. Phenotypic and functional comparison of cultures of marrow-derived mesenchymal stem cells (MSCs) and stromal cells. J Cell Physiol. 1998. 176:57–66.27. Kim DH, Yoo KH, Choi KS, Choi J, Choi SY, Yang SE, Yang YS, Im YS, Im HJ, Kim KH, Jung HL, Sung KW, Koo HH. Gene expression profile of cytokine and growth factor during differentiation of bone marrow-derived mesenchymal stem cell. Cytokine. 2005. 31:119–126.28. Bulfone-Paus S, Bulanova E, Pohl T, Budagian V, Durkop H, Ruckert R, Kunzendorf U, Paus R, Krause H. Death deflected: IL-15 inhibits TNF-alpha-mediated apoptosis in fibroblasts by TRAF2 recruitment to the IL-15R alpha chain. FASEB J. 1999. 13:1575–1585.29. Hedlund PB, Sutcliffe JG. Functional, molecular and pharmacological advances in 5-HT7 receptor research. Trends Pharmacol Sci. 2004. 25:481–486.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gene expression profile in mesenchymal stem cells derived from dental tissues and bone marrow
- Effects of Trichostatin A on the Chondrogenesis from Human Mesenchymal Stem Cells
- Clinical Safety and Efficacy of Autologous Bone Marrow-Derived Mesenchymal Stem Cell Transplantation in Sensorineural Hearing Loss Patients
- Concise Review: Differentiation of Human Adult Stem Cells Into Hepatocyte-like Cells In vitro
- Non-contact Coculture Reveals a Comprehensive Response of Chondrocytes Induced by Mesenchymal Stem Cells Through Trophic Secretion