J Vet Sci.
2010 Sep;11(3):243-247. 10.4142/jvs.2010.11.3.243.
Sequence and phylogenetic analysis of the non-structural 3A and 3B protein-coding regions of foot-and-mouth disease virus subtype A Iran 05
- Affiliations
-
- 1Department of Animal Science, Faculty of Agriculture, Tarbiat Modares University, Tehran, Iran.
- 2Department of Biotechnology, Razi Vaccine & Serum Research Institute, Karaj, Iran. m.esmaelizad@rvsri.ir
- 3Department of Quality Control, Razi Vaccine & Serum Research Institute, Karaj, Iran.
- 4Department of Animal Biotechnology, National Institute for Genetic Engineering and Biotechnology, Tehran, Iran.
- KMID: 1093499
- DOI: http://doi.org/10.4142/jvs.2010.11.3.243
Abstract
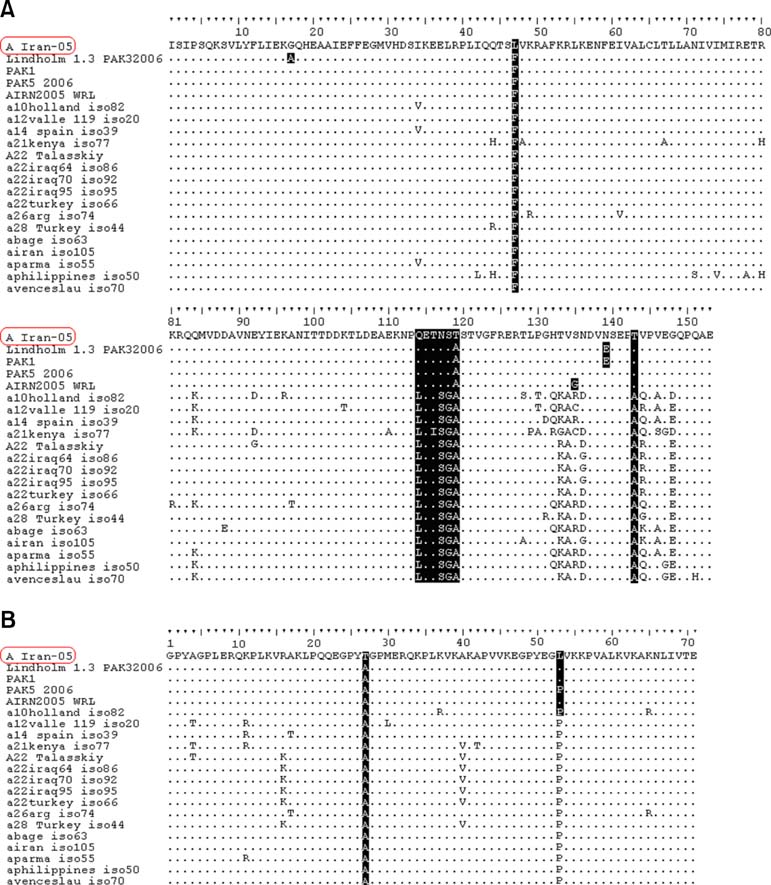
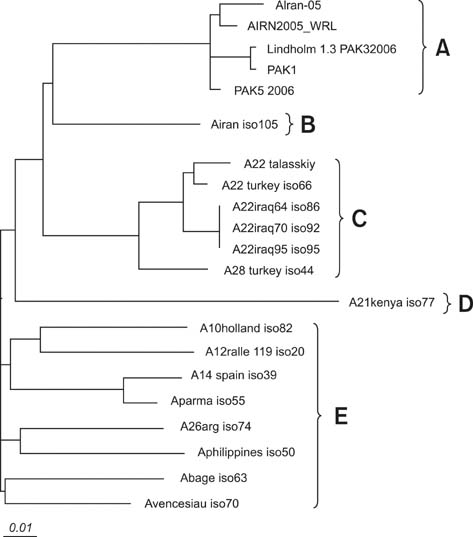
- The A Iran 05 foot-and-mouth disease virus (FMDV) subtype was detected in Iran during 2005 and has proven to be highly virulent. This study was undertaken to focus on molecular and phylogenetic analysis of 3A and 3B coding-regions in the A Iran 05 field isolate. To assess the genetic relatedness of A Iran 05 isolate the nucleotide and predicted amino acid sequences of the 3AB region of type A FMDV isolates were compared with twenty previously described type A FMDV isolates. The phylogenetic tree based on the 672 bp 3AB gene sequences of type A FMDV from thirteen different locations clustered them into five distinct lineages. The A Iran 05 isolate clustered in lineage A along with four type A variants and was closely matched with viruses isolated in Turkey and Pakistan during 2005~2006. The number of protein sequence differences exhibited by each of the isolates revealed that A Iran 05 isolate contains three amino acid substitutions at positions 47 and 119 of 3A and 27 of the 3B coding region. The nucleotide identity between A Iran 05 and the other four isolates of lineage A was estimated to be 98%.
MeSH Terms
-
Amino Acid Sequence
Base Sequence
Cluster Analysis
DNA Primers/genetics
Foot-and-Mouth Disease Virus/*genetics
Iran
Molecular Sequence Data
*Phylogeny
Reverse Transcriptase Polymerase Chain Reaction
Sequence Alignment
Sequence Analysis, DNA
Sequence Homology
Species Specificity
Viral Nonstructural Proteins/*genetics
Figure
Reference
-
1. Araújo JP Jr, Montassier HJ, Pinto AA. Extensive antigenic and genetic variation among foot-and-mouth disease type A viruses isolated from the 1994 and 1995 foci in São Paulo, Brazil. Vet Microbiol. 2002. 84:15–27.
Article2. Biswas S, Sanyal A, Hemadri D, Tosh C, Mohapatra JK, Manoj Kumar R, Bandyopadhyay SK. Sequence analysis of the non-structural 3A and 3C protein-coding regions of foot-and-mouth disease virus serotype Asia1 field isolates from an endemic country. Vet Microbiol. 2006. 116:187–193.
Article3. Curry S, Roqué-Rosell N, Zunszain PA, Leatherbarrow RJ. Foot-and-mouth disease virus 3C protease: recent structural and functional insights into an antiviral target. Int J Biochem Cell Biol. 2007. 39:1–6.
Article4. Domingo E, Baranowski E, Escarmís C, Sobrino F. Foot-and-mouth disease virus. Comp Immunol Microbiol Infect Dis. 2002. 25:297–308.
Article5. Dopazo J, Sobrino F, Palma EL, Domingo E, Moya A. Gene encoding capsid protein VP1 of foot-and-mouth disease virus: a quasispecies model of molecular evolution. Proc Natl Acad Sci USA. 1988. 85:6811–6815.
Article6. Du J, Chang H, Cong G, Shao J, Lin T, Shang Y, Liu Z, Liu X, Cai X, Xie Q. Complete nucleotide sequence of a Chinese serotype Asia1 vaccine strain of foot-and-mouth disease virus. Virus Genes. 2007. 35:635–642.
Article7. Feng Q, Yu H, Liu Y, He C, Hu J, Sang H, Ding N, Ding M, Fung YWW, Lau LT, Yu ACH, Chen J. Genome comparison of a novel foot-and-mouth disease virus with other FMDV strains. Biochem Biophys Res Commun. 2004. 323:254–263.
Article8. Gerner W, Denyer MS, Takamatsu HH, Wileman TE, Wiesmüller KH, Pfaff E, Saalmüller A. Identification of novel foot-and-mouth disease virus specific T-cell epitopes in c/c and d/d haplotype miniature swine. Virus Res. 2006. 121:223–228.
Article9. Grubman MJ. Development of novel strategies to control foot-and-mouth disease: marker vaccines and antivirals. Biologicals. 2005. 33:227–234.
Article10. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999. 41:95–98.11. Haydon DT, Bastos AD, Knowles NJ, Samuel AR. Evidence for Positive Selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics. 2001. 157:7–15.
Article12. Kamstrup S, Frimann TH, Barfoed AM. Protection of Balb/c mice against infection with FMDV by immunostimulation with CpG oligonucleotides. Antiviral Res. 2006. 72:42–48.
Article13. Klein J, Hussain M, Ahmad M, Normann P, Afzal M, Alexandersen S. Genetic characterisation of the recent foot-and-mouth disease virus subtype A/IRN/2005. Virol J. 2007. 4:122.
Article14. OIE. Situation of Foot and Mouth Disease (FMD) in the Middle East 2008-2009. 2009. Paris: OIE.15. Pacheco JM, Henry TM, O'Donnell VK, Gregory JB, Mason PW. Role of nonstructural proteins 3A and 3B in host range and pathogenicity of foot-and-mouth disease virus. J Virol. 2003. 77:13017–13027.
Article16. Perez AM, Thurmond MC, Grant PW, Carpenter TE. Use of the scan statistic on disaggregated province-based data: foot-and-mouth disease in Iran. Prev Vet Med. 2005. 71:197–207.
Article17. Sangar DV, Bryant J, Harris TJR, Brown F, Rowlands DJ. Removal of the Genome-Linked Protein of foot-and-mouth disease virus by rabbit reticulocyte lysate. J Virol. 1981. 39:67–74.
Article18. Sellers R, Gloster J. Foot-and-mouth disease: A review of intranasal infection of cattle, sheep and pigs. Vet J. 2008. 177:159–168.
Article19. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997. 24:4876–4882.
Article20. Wernery U, Kaaden OR. Foot-and-mouth disease in camelids: a review. Vet J. 2004. 168:134–142.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular characterization of a 13-amino acid deletion in VP1 (1D) protein and novel amino acid substitutions in 3D polymerase protein of foot and mouth disease virus subtype A/Iran87
- Novel foot-and-mouth disease virus in Korea, July-August 2014
- Synthetic and Adenovirus Delivered Small Interference RNA Pools Targeting Conserved Regions of Foot-and-Mouth Disease Virus
- Novel pan-lineage VP1 specific degenerate primers for precise genetic characterization of serotype O foot and mouth disease virus circulating in India
- Identification and antigenic site analysis of foot-and-mouth disease virus from pigs and cattle in Korea