Blood Res.
2015 Mar;50(1):58-61. 10.5045/br.2015.50.1.58.
Chronic eosinophilic leukemia with a FIP1L1-PDGFRA rearrangement: Two case reports and a review of Korean cases
- Affiliations
-
- 1Department of Laboratory Medicine & Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. heejinkim@skku.edu
- 2Department of Internal Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 1787916
- DOI: http://doi.org/10.5045/br.2015.50.1.58
Abstract
- No abstract available.
MeSH Terms
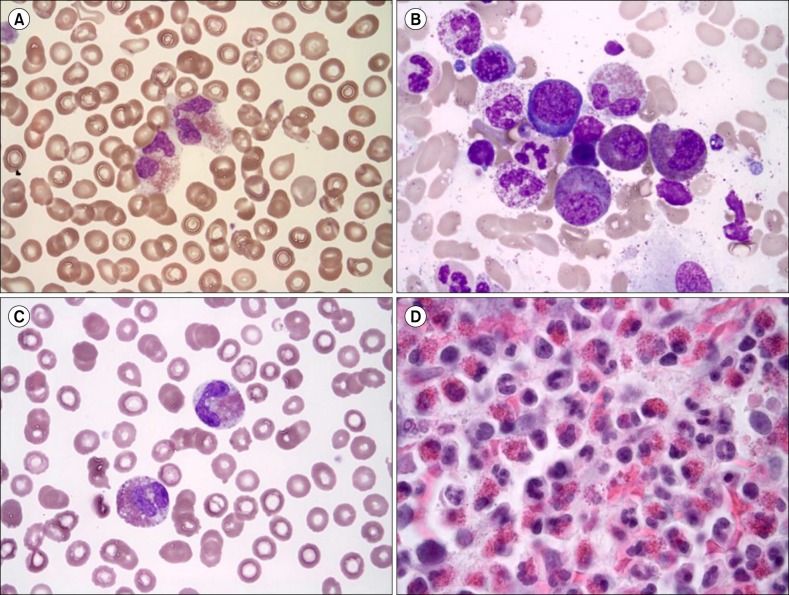
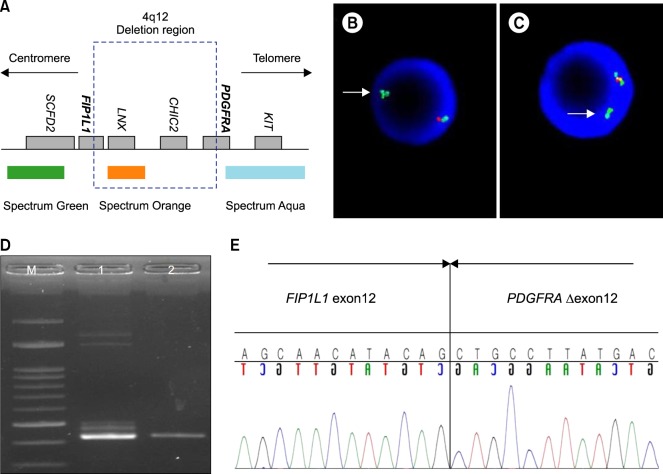
Figure
Reference
-
1. Swerdlow SH, Campo E, Harris NL, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon, France: IARC Press;2008.2. Pardanani A, Ketterling RP, Brockman SR, et al. CHIC2 deletion, a surrogate for FIP1L1-PDGFRA fusion, occurs in systemic mastocytosis associated with eosinophilia and predicts response to imatinib mesylate therapy. Blood. 2003; 102:3093–3096. PMID: 12842979.
Article3. Gotlib J, Cools J. Five years since the discovery of FIP1L1-PDGFRA: what we have learned about the fusion and other molecularly defined eosinophilias. Leukemia. 2008; 22:1999–2010. PMID: 18843283.
Article4. Kim DW, Shin MG, Yun HK, et al. Incidence and causes of hypereosinophilia (corrected) in the patients of a university hospital. Korean J Lab Med. 2009; 29:185–193. PMID: 19571614.5. Park TS, Song J, Kim JS, et al. 8p11 myeloproliferative syndrome preceded by t(8;9)(p11;q33), CEP110/FGFR1 fusion transcript: morphologic, molecular, and cytogenetic characterization of myeloid neoplasms associated with eosinophilia and FGFR1 abnormality. Cancer Genet Cytogenet. 2008; 181:93–99. PMID: 18295660.
Article6. Kim M, Lim J, Lee A, et al. A case of chronic myelomonocytic leukemia with severe eosinophilia having t(5;12)(q31;p13) with t(1;7)(q10;p10). Acta Haematol. 2005; 114:104–107. PMID: 16103634.
Article7. Lee H, Kim M, Lim J, et al. Acute myeloid leukemia associated with FGFR1 abnormalities. Int J Hematol. 2013; 97:808–812. PMID: 23609419.
Article8. Jang SE, Kang HJ, Chang YH, et al. A case of myeloid neoplasm with the PDGFRB rearrangement and eosinophilia. Korean J Med. 2010; 78:386–390.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Loeffler endocarditis in chronic eosinophilic leukemia with FIP1L1/PDGFRA rearrangement: full recovery with low dose imatinib
- Chronic eosinophilic leukemia with a FIP1L1-PDGFRA rearrangement: Two case reports and a review of Korean cases
- Chronic eosinophilic leukemia with FIP1L1-PDGFRA rearrangement
- A Case of Atypical Chronic Myeloid Leukemia with the JAK2V617F Mutation
- detection of BCR-ABL gene rearrangement by use of the polymerase chain reaction in chronic myeloid leukemia