J Bone Metab.
2019 Feb;26(1):3-12. 10.11005/jbm.2019.26.1.3.
The Function of the Vitamin D Receptor and a Possible Role of Enhancer RNA in Epigenomic Regulation of Target Genes: Implications for Bone Metabolism
- Affiliations
-
- 1Department of Molecular Endocrinology, Fujii Memorial Institute of Medical Sciences, Institute of Advanced Medical Sciences, Tokushima University, Tokushima, Japan.
- 2Center for Regional Cooperation, Iwaki Meisei University, Iwaki, Japan. shigeaki.kato@iwakimu.ac.jp, uskato0525@gmail.com
- 3Research Institute of Innovative Medicine, Tokiwa Foundation, Jyoban Kamiyunagayamachi, Iwaki, Japan.
- KMID: 2440639
- DOI: http://doi.org/10.11005/jbm.2019.26.1.3
Abstract
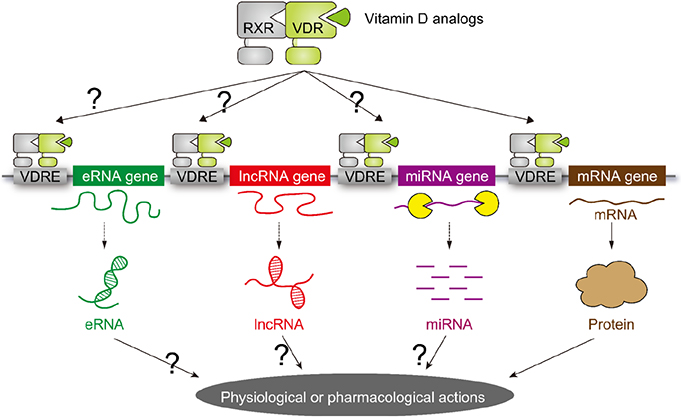
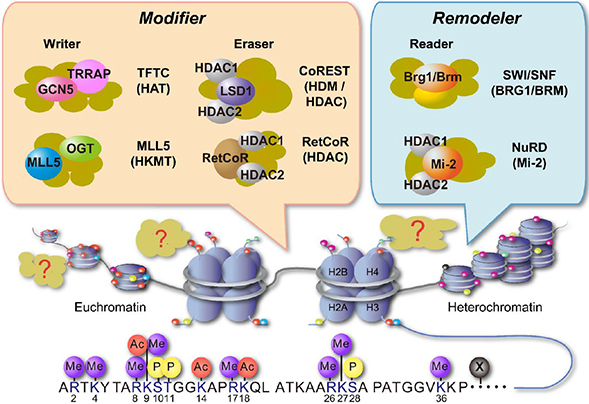
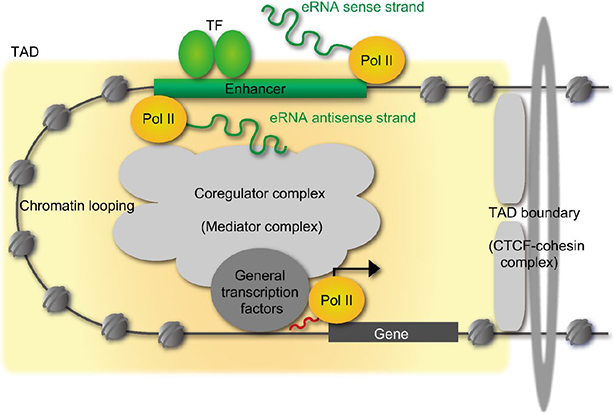
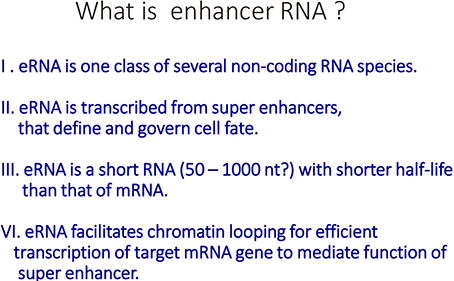
- Vitamin D (VD) is essential for bone health, and VD or its analogues are widely used in clinics to ameliorate bone loss. The targets and mode of VD anti-osteoporotic actions appear to be different from those of other classes of drugs modulating bone remodeling. VD exerts its biological activities through the nuclear VD receptor (VDR)-mediated transcriptional regulation of target mRNA and non-coding RNA genes. VD-induced gene regulation involves epigenetic modifications of chromatin conformation at the target loci as well as reconfiguration of higher-order chromosomal organization through VDR-mediated recruitment of various regulatory factors. Enhancer RNAs (eRNA), a class of non-coding enhancer-derived RNAs, have recently emerged as VDR target gene candidates that act through reorganization of chromatin looping to induce enhancer-promoter interaction in activation of mRNA-encoding genes. This review outlines the molecular mechanisms of VD actions mediated by the VDR and suggests novel function of eRNAs in VDR transactivation.
MeSH Terms
Figure
Reference
-
1. Morris HA, Anderson PH. Autocrine and paracrine actions of vitamin D. Clin Biochem Rev. 2010; 31:129–138.2. Feldman D, Malloy PJ. Mutations in the vitamin D receptor and hereditary vitamin D-resistant rickets. Bonekey Rep. 2014; 3:510.
Article3. Kato S. The function of vitamin D receptor in vitamin D action. J Biochem. 2000; 127:717–722.
Article4. Haussler MR, Whitfield GK, Haussler CA, et al. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998; 13:325–349.
Article5. Matsumoto T, Ito M, Hayashi Y, et al. A new active vitamin D3 analog, eldecalcitol, prevents the risk of osteoporotic fractures-a randomized, active comparator, double-blind study. Bone. 2011; 49:605–612.
Article6. Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461.
Article7. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012; 489:101–108.8. Takeyama K, Kitanaka S, Sato T, et al. 25-Hydroxyvitamin D3 1alpha-hydroxylase and vitamin D synthesis. Science. 1997; 277:1827–1830.
Article9. Kitanaka S, Takeyama K, Murayama A, et al. Inactivating mutations in the 25-hydroxyvitamin D3 1alpha-hydroxylase gene in patients with pseudovitamin D-deficiency rickets. N Engl J Med. 1998; 338:653–661.
Article10. Yoshizawa T, Handa Y, Uematsu Y, et al. Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat Genet. 1997; 16:391–396.
Article11. Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995; 83:835–839.
Article12. Pike JW. Genome-wide principles of gene regulation by the vitamin D receptor and its activating ligand. Mol Cell Endocrinol. 2011; 347:3–10.
Article13. Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008; 132:958–970.
Article14. Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016; 37:76–81.
Article15. Seuter S, Neme A, Carlberg C. Epigenomic PU.1-VDR crosstalk modulates vitamin D signaling. Biochim Biophys Acta Gene Regul Mech. 2017; 1860:405–415.
Article16. Rachez C, Suldan Z, Ward J, et al. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998; 12:1787–1800.
Article17. Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006; 20:1405–1428.
Article18. Kato S, Yokoyama A, Fujiki R. Nuclear receptor coregulators merge transcriptional coregulation with epigenetic regulation. Trends Biochem Sci. 2011; 36:272–281.
Article19. Seuter S, Heikkinen S, Carlberg C. Chromatin acetylation at transcription start sites and vitamin D receptor binding regions relates to effects of 1alpha,25-dihydroxyvitamin D3 and histone deacetylase inhibitors on gene expression. Nucleic Acids Res. 2013; 41:110–124.
Article20. Battaglia S, Karasik E, Gillard B, et al. LSD1 dual function in mediating epigenetic corruption of the vitamin D signaling in prostate cancer. Clin Epigenetics. 2017; 9:82.
Article21. Masuyama R, Nakaya Y, Tanaka S, et al. Dietary phosphorus restriction reverses the impaired bone mineralization in vitamin D receptor knockout mice. Endocrinology. 2001; 142:494–497.
Article22. Yamamoto Y, Yoshizawa T, Fukuda T, et al. Vitamin D receptor in osteoblasts is a negative regulator of bone mass control. Endocrinology. 2013; 154:1008–1020.
Article23. Uenishi K, Tokiwa M, Kato S, et al. Stimulation of intestinal calcium absorption by orally administrated vitamin D3 compounds: a prospective open-label randomized trial in osteoporosis. Osteoporos Int. 2018; 29:723–732.
Article24. Bouillon R, Carmeliet G, Verlinden L, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008; 29:726–776.
Article25. Lieben L, Masuyama R, Torrekens S, et al. Normocalcemia is maintained in mice under conditions of calcium malabsorption by vitamin D-induced inhibition of bone mineralization. J Clin Invest. 2012; 122:1803–1815.
Article26. Nakamura T, Takano T, Fukunaga M, et al. Eldecalcitol is more effective for the prevention of osteoporotic fractures than alfacalcidol. J Bone Miner Metab. 2013; 31:417–422.
Article27. Sakai Y, Kishimoto J, Demay MB. Metabolic and cellular analysis of alopecia in vitamin D receptor knockout mice. J Clin Invest. 2001; 107:961–966.
Article28. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011; 12:861–874.
Article29. Meyer MB, Benkusky NA, Lee CH, et al. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J Biol Chem. 2014; 289:19539–19554.
Article30. Seuter S, Pehkonen P, Heikkinen S, et al. Dynamics of 1alpha, 25-dihydroxyvitamin D3-dependent chromatin accessibility of early vitamin D receptor target genes. Biochim Biophys Acta. 2013; 1829:1266–1275.
Article31. Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012; 13:436–447.
Article32. Allis CD, Berger SL, Cote J, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007; 131:633–636.
Article33. Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011; 80:473–499.
Article34. Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011; 146:1016–1028.
Article35. Fujiki R, Hashiba W, Sekine H, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011; 480:557–560.
Article36. Baba A, Ohtake F, Okuno Y, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 2011; 13:668–675.
Article37. Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012; 13:343–357.
Article38. Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014; 15:93–106.
Article39. Kato S, Ishii T, Kouzmenko A. Point mutations in an epigenetic factor lead to multiple types of bone tumors: role of H3.3 histone variant in bone development and disease. Bonekey Rep. 2015; 4:715.
Article40. Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013; 20:14–22.
Article41. Sawatsubashi S, Murata T, Lim J, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010; 24:159–170.
Article42. Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005; 437:436–439.
Article43. Ren B, Dixon JR. A CRISPR connection between chromatin topology and genetic disorders. Cell. 2015; 161:955–957.
Article44. Hnisz D, Shrinivas K, Young RA, et al. A phase separation model for transcriptional control. Cell. 2017; 169:13–23.
Article45. Kim TK, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015; 162:948–959.
Article46. Neme A, Seuter S, Carlberg C. Vitamin D-dependent chromatin association of CTCF in human monocytes. Biochim Biophys Acta. 2016; 1859:1380–1388.
Article47. Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016; 17:207–223.
Article48. Li W, Notani D, Ma Q, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013; 498:516–520.
Article49. Lam MT, Cho H, Lesch HP, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013; 498:511–515.
Article50. Sawatsubashi S, Joko Y, Fukumoto S, et al. Development of versatile non-homologous end joining-based knock-in module for genome editing. Sci Rep. 2018; 8:593.
Article51. Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010; 285:15599–15610.
Article52. Tsai PF, Dell'Orso S, Rodriguez J, et al. A muscle-specific enhancer RNA mediates cohesin recruitment and regulates transcription in trans. Mol Cell. 2018; 71:129–141.
Article53. Weintraub AS, Li CH, Zamudio AV, et al. YY1 Is a structural regulator of enhancer-promoter loops. Cell. 2017; 171:1573–1588.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gestational Diabetes Mellitus, Fetal Growth and Vitamin D
- Vitamin D regulation of adipogenesis and adipose tissue functions
- Vitamin D Activities for Health Outcomes
- Gene Expression Regulation by Agonist-Independent Constitutive Signaling of Melanocortin-1 Receptor
- The Association between Vitamin D Deficiency and Perinatal Outcomes of Pregnancy