J Korean Med Assoc.
2015 Nov;58(11):995-1002. 10.5124/jkma.2015.58.11.995.
Non-invasive prenatal test using cell free DNA
- Affiliations
-
- 1Hamchoon Women's Clinic, Seoul, Korea. mdkkw@hamchoon.com
- KMID: 2195128
- DOI: http://doi.org/10.5124/jkma.2015.58.11.995
Abstract
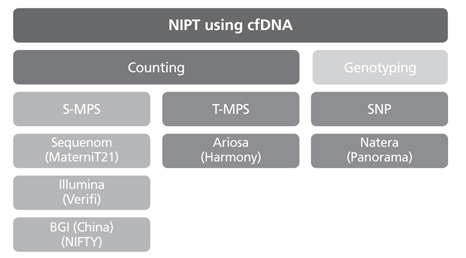
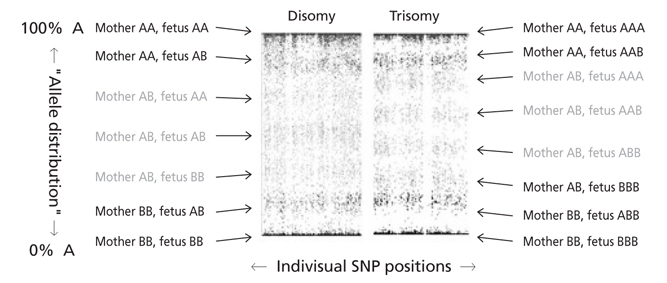
- Although conventional prenatal screening tests for Down syndrome have been developed over the past 20 years, the positive predictive value of these tests is around 5%. Through these tests, many pregnant women have taken invasive tests including chorionic villi sampling and amniocentesis for confirming Down syndrome. Invasive test carries the risk of fetal loss at a low but significant rate. There is a large amount of evidence that non-invasive prenatal test (NIPT) using cell free DNA in maternal serum is more sensitive and specific than conventional maternal serum and/or ultrasound screening. Therefore implementing NIPT will increase aneuploidy detection rate and concurrently decrease fetal loss rate accompanying invasive test. More than 1,000,000 NIPT were performed globally since 2011. The uptake rate of NIPT is expected to increase more rapidly in the future. Moreover, as a molecular genetic technique advances, NIPT can be used for not only common aneuploidy screening but single gene disorder, microdeletion, and whole fetal genome sequencing. In this review, I will focus on the NIPT for common aneuploidies such as trisomy 13, 18, and 21.
MeSH Terms
Figure
Cited by 1 articles
-
Shift of paradigm in prenatal diagnosis
Do Yeong Hwang
J Korean Med Assoc. 2015;58(11):976-978. doi: 10.5124/jkma.2015.58.11.976.
Reference
-
1. Weijerman ME, de Winter JP. Clinical practice: the care of children with Down syndrome. Eur J Pediatr. 2010; 169:1445–1452.2. Nicolaides KH. Screening for fetal aneuploidies at 11 to 13 weeks. Prenat Diagn. 2011; 31:7–15.
Article3. Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997; 350:485–487.
Article4. Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004; 50:88–92.
Article5. Lun FM, Chiu RW, Chan KC, Leung TY, Lau TK, Lo YM. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008; 54:1664–1672.
Article6. Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem. 2010; 56:1279–1286.
Article7. Nygren AO, Dean J, Jensen TJ, Kruse S, Kwong W, van den Boom D, Ehrich M. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin Chem. 2010; 56:1627–1635.
Article8. Sikora A, Zimmermann BG, Rusterholz C, Birri D, Kolla V, Lapaire O, Hoesli I, Kiefer V, Jackson L, Hahn S. Detection of increased amounts of cell-free fetal DNA with short PCR amplicons. Clin Chem. 2010; 56:136–138.
Article9. Nicolaides KH, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am J Obstet Gynecol. 2012; 207:374.e1–374.e6.
Article10. Ashoor G, Poon L, Syngelaki A, Mosimann B, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks' gestation: effect of maternal and fetal factors. Fetal Diagn Ther. 2012; 31:237–243.
Article11. Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999; 64:218–224.
Article12. Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, Gammill HS, Rubens CE, Santillan DA, Murray JC, Tabor HK, Bamshad MJ, Eichler EE, Shendure J. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012; 4:137ra76.
Article13. Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR. Non-invasive prenatal measurement of the fetal genome. Nature. 2012; 487:320–324.
Article14. Lun FM, Chiu RW, Sun K, Leung TY, Jiang P, Chan KC, Sun H, Lo YM. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin Chem. 2013; 59:1583–1594.
Article15. Tsui NB, Jiang P, Wong YF, Leung TY, Chan KC, Chiu RW, Sun H, Lo YM. Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem. 2014; 60:954–962.
Article16. Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, Foo CH, Xie B, Tsui NB, Lun FM, Zee BC, Lau TK, Cantor CR, Lo YM. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008; 105:20458–20463.
Article17. Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008; 105:16266–16271.
Article18. Sparks AB, Wang ET, Struble CA, Barrett W, Stokowski R, McBride C, Zahn J, Lee K, Shen N, Doshi J, Sun M, Garrison J, Sandler J, Hollemon D, Pattee P, Tomita-Mitchell A, Mitchell M, Stuelpnagel J, Song K, Oliphant A. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn. 2012; 32:3–9.
Article19. Zimmermann B, Hill M, Gemelos G, Demko Z, Banjevic M, Baner J, Ryan A, Sigurjonsson S, Chopra N, Dodd M, Levy B, Rabinowitz M. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat Diagn. 2012; 32:1233–1241.
Article20. Hall MP, Hill M, Zimmermann B, Sigurjonsson S, Westemeyer M, Saucier J, Demko Z, Rabinowitz M. Non-invasive prenatal detection of trisomy 13 using a single nucleotide polymorphism- and informatics-based approach. PLoS One. 2014; 9:e96677.
Article21. Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2015; 45:249–266.
Article22. Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, Tomlinson MW, Pereira L, Spitz JL, Hollemon D, Cuckle H, Musci TJ, Wapner RJ. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015; 372:1589–1597.
Article23. Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Grody WW, Nelson SF, Canick JA. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012; 14:296–305.
Article24. Kalousek DK, Vekemans M. Confined placental mosaicism. J Med Genet. 1996; 33:529–533.
Article25. Choi H, Lau TK, Jiang FM, Chan MK, Zhang HY, Lo PS, Chen F, Zhang L, Wang W. Fetal aneuploidy screening by maternal plasma DNA sequencing: 'false positive' due to confined placental mosaicism. Prenat Diagn. 2013; 33:198–200.
Article26. Futch T, Spinosa J, Bhatt S, de Feo E, Rava RP, Sehnert AJ. Initial clinical laboratory experience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat Diagn. 2013; 33:569–574.
Article27. Russell LM, Strike P, Browne CE, Jacobs PA. X chromosome loss and ageing. Cytogenet Genome Res. 2007; 116:181–185.
Article28. Wang Y, Chen Y, Tian F, Zhang J, Song Z, Wu Y, Han X, Hu W, Ma D, Cram D, Cheng W. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin Chem. 2014; 60:251–259.
Article29. Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, Garber JE, Wilkins-Haug L, Vora NL, Warsof S, Goldberg J, Ziainia T, Halks-Miller M. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015; 314:162–169.
Article30. Snyder MW, Simmons LE, Kitzman JO, Coe BP, Henson JM, Daza RM, Eichler EE, Shendure J, Gammill HS. Copy-number variation and false positive prenatal aneuploidy screening results. N Engl J Med. 2015; 372:1639–1645.
Article31. Smith M, Lewis KM, Holmes A, Visootsak J. A case of false negative NIPT for Down syndrome-lessons learned. Case Rep Genet. 2014; 2014:823504.
Article32. Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. 2013; 33:667–674.
Article33. Palomaki GE, Kloza EM, Lambert-Messerlian GM, van den Boom D, Ehrich M, Deciu C, Bombard AT, Haddow JE. Circulating cell free DNA testing: are some test failures informative? Prenat Diagn. 2015; 35:289–293.
Article34. Dar P, Curnow KJ, Gross SJ, Hall MP, Stosic M, Demko Z, Zimmermann B, Hill M, Sigurjonsson S, Ryan A, Banjevic M, Kolacki PL, Koch SW, Strom CM, Rabinowitz M, Benn P. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based noninvasive prenatal aneuploidy testing. Am J Obstet Gynecol. 2014; 211:527.e1.
Article35. Wegrzyn P, Faro C, Falcon O, Peralta CF, Nicolaides KH. Placental volume measured by three-dimensional ultrasound at 11 to 13+6 weeks of gestation: relation to chromosomal defects. Ultrasound Obstet Gynecol. 2005; 26:28–32.
Article36. Norton ME, Jelliffe-Pawlowski LL, Currier RJ. Chromosome abnormalities detected by current prenatal screening and noninvasive prenatal testing. Obstet Gynecol. 2014; 124:979–986.
Article37. Petersen OB, Vogel I, Ekelund C, Hyett J, Tabor A. Danish Fetal Medicine Study Group. Danish Clinical Genetics Study Group. Potential diagnostic consequences of applying non-invasive prenatal testing: population-based study from a country with existing first-trimester screening. Ultrasound Obstet Gynecol. 2014; 43:265–271.
Article38. Leung TY, Qu JZ, Liao GJ, Jiang P, Cheng YK, Chan KC, Chiu RW, Lo YM. Noninvasive twin zygosity assessment and aneuploidy detection by maternal plasma DNA sequencing. Prenat Diagn. 2013; 33:675–681.
Article39. Lau TK, Jiang F, Chan MK, Zhang H, Lo PS, Wang W. Noninvasive prenatal screening of fetal Down syndrome by maternal plasma DNA sequencing in twin pregnancies. J Matern Fetal Neonatal Med. 2013; 26:434–437.
Article40. Huang X, Zheng J, Chen M, Zhao Y, Zhang C, Liu L, Xie W, Shi S, Wei Y, Lei D, Xu C, Wu Q, Guo X, Shi X, Zhou Y, Liu Q, Gao Y, Jiang F, Zhang H, Su F, Ge H, Li X, Pan X, Chen S, Chen F, Fang Q, Jiang H, Lau TK, Wang W. Noninvasive prenatal testing of trisomies 21 and 18 by massively parallel sequencing of maternal plasma DNA in twin pregnancies. Prenat Diagn. 2014; 34:335–340.
Article41. del Mar Gil M, Quezada MS, Bregant B, Syngelaki A, Nicolaides KH. Cell-free DNA analysis for trisomy risk assessment in first-trimester twin pregnancies. Fetal Diagn Ther. 2014; 35:204–211.
Article42. Committee Opinion No. 640: cell-free DNA screening for fetal aneuploidy. Obstet Gynecol. 2015; 126:e31–e37.43. American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 545: noninvasive prenatal testing for fetal aneuploidy. Obstet Gynecol. 2012; 120:1532–1534.44. Cuckle H, Benn P, Pergament E. Cell-free DNA screening for fetal aneuploidy as a clinical service. Clin Biochem. 2015; 48:932–941.
Article45. Benn P, Borrell A, Chiu RW, Cuckle H, Dugoff L, Faas B, Gross S, Huang T, Johnson J, Maymon R, Norton M, Odibo A, Schielen P, Spencer K, Wright D, Yaron Y. Position statement from the Chromosome Abnormality Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn. 2015; 35:725–734.
Article46. Hill M, Wright D, Daley R, Lewis C, McKay F, Mason S, Lench N, Howarth A, Boustred C, Lo K, Plagnol V, Spencer K, Fisher J, Kroese M, Morris S, Chitty LS. Evaluation of non-invasive prenatal testing (NIPT) for aneuploidy in an NHS setting: a reliable accurate prenatal non-invasive diagnosis (RAPID) protocol. BMC Pregnancy Childbirth. 2014; 14:229.
Article47. Langlois S, Brock JA. Genetics Committee. Wilson RD, Audibert F, Brock JA, Carroll J, Cartier L, Gagnon A, Johnson JA, Langlois S, Macdonald W, Murphy-Kaulbeck L, Okun N, Pastuck M, Senikas V. Current status in non-invasive prenatal detection of Down syndrome, trisomy 18, and trisomy 13 using cell-free DNA in maternal plasma. J Obstet Gynaecol Can. 2013; 35:177–183.
Article48. Dondorp W, de Wert G, Bombard Y, Bianchi DW, Bergmann C, Borry P, Chitty LS, Fellmann F, Forzano F, Hall A, Henneman L, Howard HC, Lucassen A, Ormond K, Peterlin B, Radojkovic D, Rogowski W, Soller M, Tibben A, Tranebjærg L, van El CG, Cornel MC. Non-invasive prenatal testing for aneuploidy and beyond: challenges of responsible innovation in prenatal screening. Eur J Hum Genet. 2015; 23:1438–1450.
Article49. Wilson KL, Czerwinski JL, Hoskovec JM, Noblin SJ, Sullivan CM, Harbison A, Campion MW, Devary K, Devers P, Singletary CN. NSGC practice guideline: prenatal screening and diagnostic testing options for chromosome aneuploidy. J Genet Couns. 2013; 22:4–15.
Article50. Salomon LJ, Alfirevic Z, Bilardo CM, Chalouhi GE, Ghi T, Kagan KO, Lau TK, Papageorghiou AT, Raine-Fenning NJ, Stirnemann J, Suresh S, Tabor A, Timor-Tritsch IE, Toi A, Yeo G. ISUOG practice guidelines: performance of first-trimester fetal ultrasound scan. Ultrasound Obstet Gynecol. 2013; 41:102–113.
Article51. Gregg AR, Gross SJ, Best RG, Monaghan KG, Bajaj K, Skotko BG, Thompson BH, Watson MS. ACMG statement on noninvasive prenatal screening for fetal aneuploidy. Genet Med. 2013; 15:395–398.
Article52. Grace MR, Hardisty E, Green NS, Davidson E, Stuebe AM, Vora NL. Cell free DNA testing-interpretation of results using an online calculator. Am J Obstet Gynecol. 2015; 213:30–32.
Article53. van den Heuvel A, Chitty L, Dormandy E, Newson A, Deans Z, Attwood S, Haynes S, Marteau TM. Will the introduction of non-invasive prenatal diagnostic testing erode informed choices? An experimental study of health care professionals. Patient Educ Couns. 2010; 78:24–28.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical application of non-invasive prenatal testing using cell free fetal DNA
- An Experience of Using the Harmony Test for Genomics-Based Non-Invasive Prenatal Testing
- Prenatal maternal serum screening for fetal aneuploidy
- Non-invasive prenatal diagnosis of fetal trisomy 21 using cell-free fetal DNA in maternal blood
- Advantages of the single nucleotide polymorphism-based noninvasive prenatal test