J Korean Soc Magn Reson Med.
2011 Aug;15(2):110-122. 10.13104/jksmrm.2011.15.2.110.
Anatomical Brain Connectivity Map of Korean Children
- Affiliations
-
- 1BK21 Project for Medical Science, Yonsei University College of Medicine, Seoul, Republic of Korea. parkhj@yuhs.ac
- 2Severance Biomedical Science Institute, Department of Diagnostic Radiology, Yonsei University College of Medicine, Seoul, Republic of Korea.
- KMID: 2000059
- DOI: http://doi.org/10.13104/jksmrm.2011.15.2.110
Abstract
- PURPOSE
The purpose of this study is to establish the method generating human brain anatomical connectivity from Korean children and evaluating the network topological properties using small-world network analysis.
MATERIALS AND METHODS
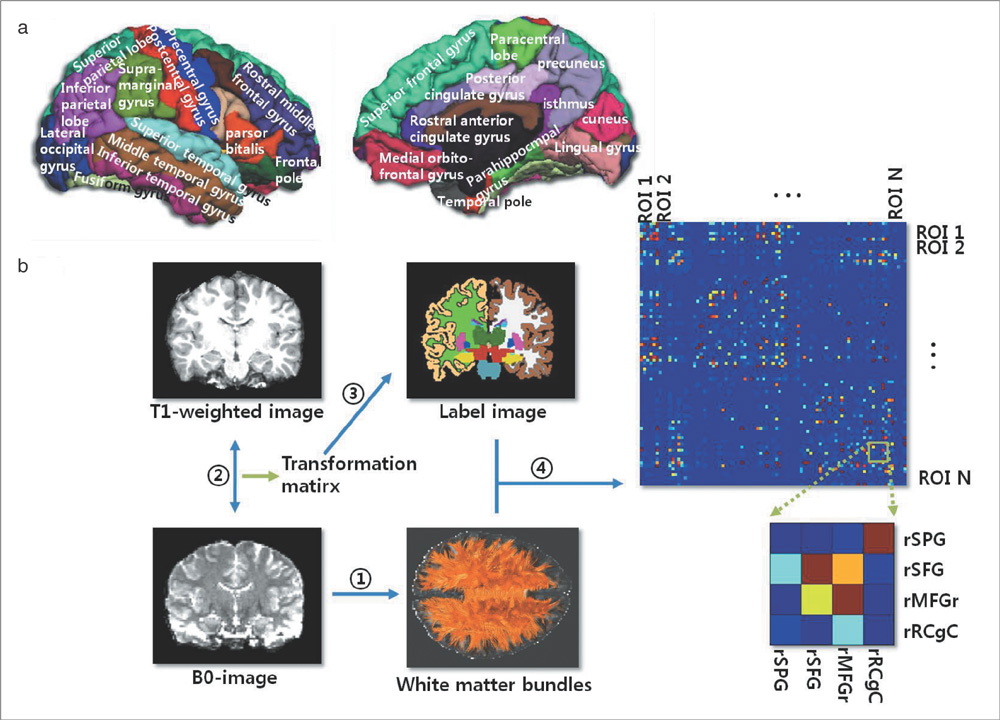
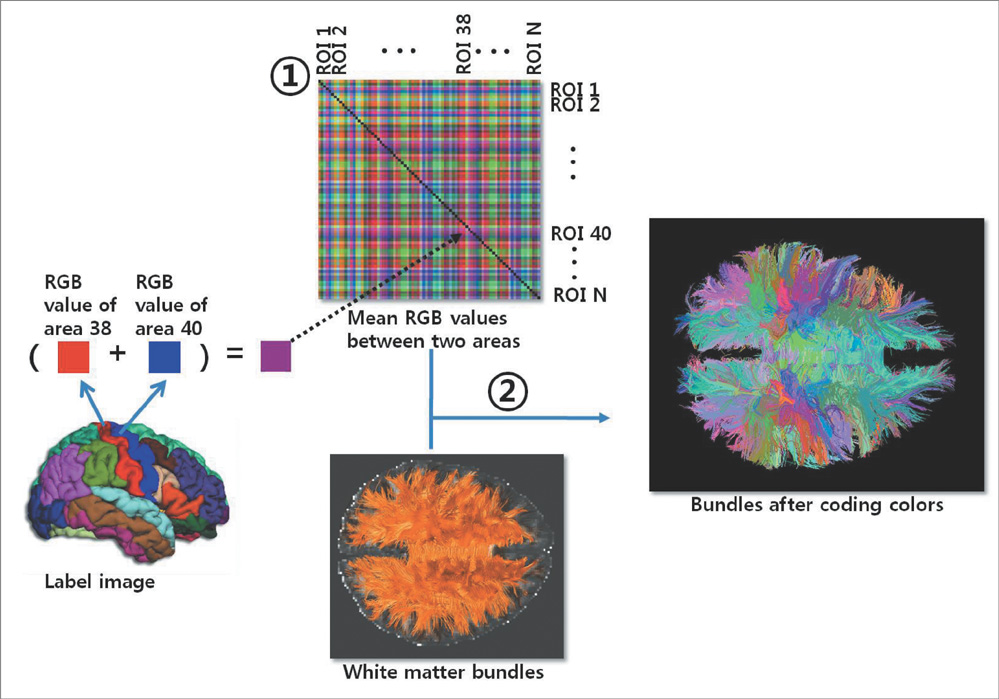
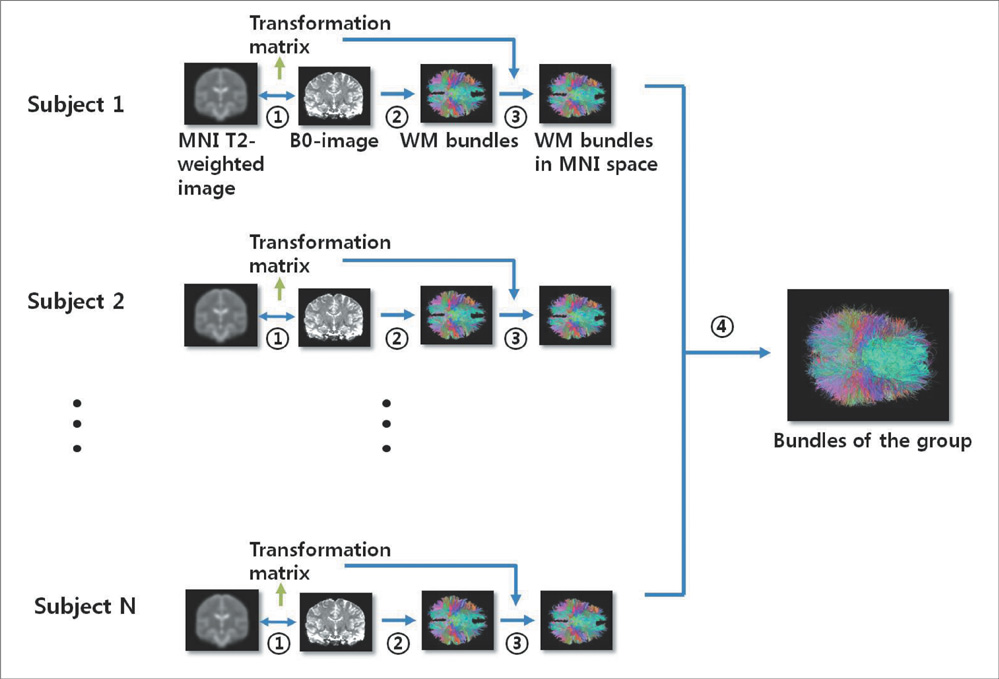
Using diffusion tensor images (DTI) and parcellation maps of structural MRIs acquired from twelve healthy Korean children, we generated a brain structural connectivity matrix for individual. We applied one sample t-test to the connectivity maps to derive a representative anatomical connectivity for the group. By spatially normalizing the white matter bundles of participants into a template standard space, we obtained the anatomical brain network model. Network properties including clustering coefficient, characteristic path length, and global/local efficiency were also calculated.
RESULTS
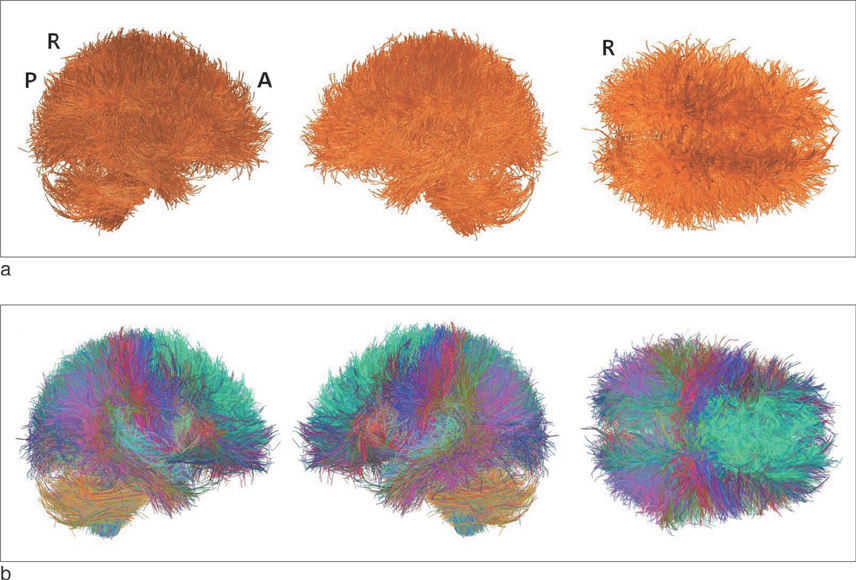
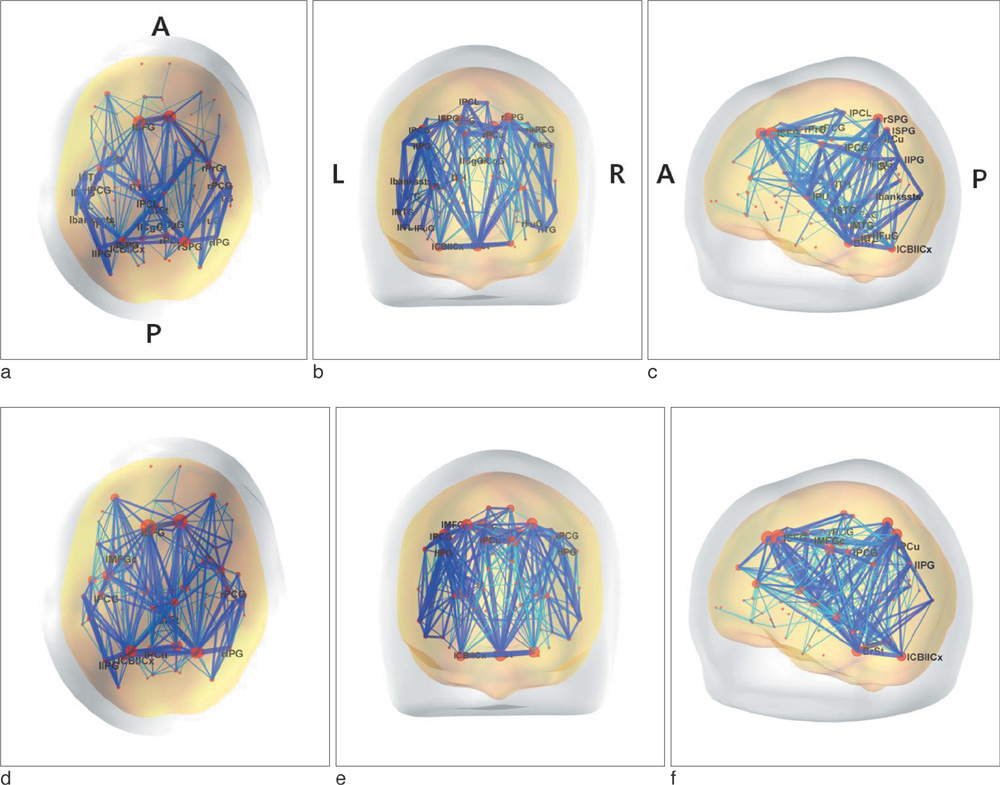
We found that the structural connectivity of Korean children group preserves the small-world properties. The anatomical connectivity map obtained in this study showed that children group had higher intra-hemispheric connectivity than inter-hemispheric connectivity. We also observed that the neural connectivity of the group is high between brain stem and motorsensory areas.
CONCLUSION
We suggested a method to examine the anatomical brain network of Korean children group. The proposed method can be used to evaluate the efficiency of anatomical brain networks in people with disease.
MeSH Terms
Figure
Reference
-
1. Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009. 10:186–198.2. Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011.3. Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J Neurosci. 2006. 26:63–72.4. Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006. 12:512–523.5. Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001. 98:676–682.6. Smit DJ, Stam CJ, Posthuma D, Boomsma DI, de Geus EJ. Heritability of "small-world" networks in the brain: a graph theoretical analysis of resting-state EEG functional connectivity. Hum Brain Mapp. 2008. 29:1368–1378.7. Valencia M, Martinerie J, Dupont S, Chavez M. Dynamic small-world behavior in functional brain networks unveiled by an event-related networks approach. Phys Rev E Stat Nonlin Soft Matter Phys. 2008. 77:050905.8. Zhou C, Zemanova L, Zamora G, Hilgetag CC, Kurths J. Hierarchical organization unveiled by functional connectivity in complex brain networks. Phys Rev Lett. 2006. 97:238103.9. Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, et al. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex. 2009. 19:524–536.10. Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009. 19:72–78.11. Eguiluz VM, Chialvo DR, Cecchi GA, Baliki M, Apkarian AV. Scale-free brain functional networks. Phys Rev Lett. 2005. 94:018102.12. Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005. 15:1332–1342.13. Ferrarini L VI, Baerends E, van Tol MJ, Renken RJ, van der Wee NJ, Veltman DJ, et al. Hierarchical functional modularity in the resting-state human brain. Hum Brain Mapp. 2009. 30:2220–2231.14. Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. Neuroimage. 2009. 44:715–723.15. Stam CJ. Functional connectivity patterns of human magnetoencephalographic recordings: a 'small-world' network? Neurosci Lett. 2004. 355:25–28.16. Achard S, Bassett DS, Meyer-Lindenberg A, Bullmore E. Fractal connectivity of long-memory networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2008. 77:036104.17. Linkenkaer-Hansen K, Nikouline VV, Palva JM, Ilmoniemi RJ. Long-Range Temporal Correlations and Scaling Behavior in Human Brain Oscillations. The Journal of Neuroscience. 2001. 21:1370–1377.18. Maxim V, Sendur L, Fadili J, Suckling J, Gould R, Howard R, et al. Fractional Gaussian noise, functional MRI and Alzheimer's disease. Neuroimage. 2005. 25:141–158.19. Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991. 1:1–47.20. Hilgetag CC, Burns GA, O'Neill MA, Scannell JW, Young MP. Anatomical connectivity defines the organization of clusters of cortical areas in the macaque monkey and the cat. Philos Trans R Soc Lond B Biol Sci. 2000. 355:91–110.21. Latora V, Marchiori M. Economic small-world behavior in weighted networks. Eur Phys J B. 2003. 32:249–263.22. Scannell JW, Burns GA, Hilgetag CC, O'Neil MA, Young MP. The connectional organization of the cortico-thalamic system of the cat. Cereb Cortex. 1999. 9:277–299.23. Young MP. Objective analysis of the topological organization of the primate cortical visual system. Nature. 1992. 358:152–155.24. Lee JD, Park HJ, Park ES, Oh MK, Park B, Rha DW, et al. Motor pathway injury in patients with periventricular leucomalacia and spastic diplegia. Brain. 2011.25. Park HJ. Quantification of white matter using diffusion-tensor imaging. Int Rev Neurobiol. 2005. 66:167–212.26. Bullmore ET, Bassett DS. Brain graphs: graphical models of the human brain connectome. Annu Rev Clin Psychol. 2011. 7:113–140.27. Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen VJ, Meuli R, Thiran JP. Mapping Human Whole-Brain Structural Networks with Diffusion MRI. PLoS ONE. 2007. 7:e597.28. Counsell SJ, Dyet LE, Larkman DJ, Nunes RG, Boardman JP, Allsop JM, et al. Thalamo-cortical connectivity in children born preterm mapped using probabilistic magnetic resonance tractography. Neuroimage. 2007. 34:896–904.29. Lenroot RK, Giedd JN. Brain development in children and adolescents: insights from anatomical magnetic resonance imaging. Neurosci Biobehav Rev. 2006. 30:718–729.30. Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, et al. The maturing architecture of the brain's default network. Proc Natl Acad Sci U S A. 2008. 105:4028–4032.31. Park HJ, Kim JJ, Lee SK, Seok JH, Chun J, Kim DI, et al. Corpus callosal connection mapping using cortical gray matter parcellation and DT-MRI. Hum Brain Mapp. 2008. 29:503–516.32. Friston KJ. Commentary and opinion: II. Statistical parametric mapping: ontology and current issues. J Cereb Blood Flow Metab. 1995. 15:361–370.33. Kim DJ, Park HJ, Kang KW, Shin YW, Kim JJ, Moon WJ, et al. How does distortion correction correlate with anisotropic indices? A diffusion tensor imaging study. Magn Reson Imaging. 2006. 24:1369–1376.34. Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999. 9:179–194.35. Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999. 9:195–207.36. Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004. 14:11–22.37. Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010. 52:1059–1069.38. Zalesky A, Fornito A, Harding IH, Cocchi L, Yucel M, Pantelis C, et al. Whole-brain anatomical networks: does the choice of nodes matter? Neuroimage. 2010. 50:970–983.39. Sporns O, Tononi G, Edelman GM. Theoretical neuroanatomy and the connectivity of the cerebral cortex. Behav Brain Res. 2002. 135:69–74.40. Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011.41. Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008. 9:417–422.42. Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 2003. 100:253–258.43. Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, et al. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009. 106:2035–2040.44. Koch MA, Norris DG, Hund-Georgiadis M. An investigation of functional and anatomical connectivity using magnetic resonance imaging. Neuroimage. 2002. 16:241–250.45. Iturria-Medina Y, Sotero RC, Canales-Rodriguez EJ, Aleman-Gomez Y, Melie-Garcia L. Studying the human brain anatomical network via diffusion-weighted MRI and Graph Theory. Neuroimage. 2008. 40:1064–1076.46. Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006. 31:968–980.47. Hofer S, Frahm J. Topography of the human corpus callosum revisited - Comprehensive fiber tractography using diffusion tensor magnetic resonance imaging. Neuroimage. 2006. 32:989–994.48. Huang H, Zhang J, Jiang H, Wakana S, Poetscher L, Miller MI, et al. DTI tractography based parcellation of white matter: application to the mid-sagittal morphology of corpus callosum. Neuroimage. 2005. 26:195–205.49. Kim M, Ronen I, Ugurbil K, Kim DS. Spatial resolution dependence of DTI tractography in human occipito-callosal region. Neuroimage. 2006. 32:1243–1249.50. Catani M, Thiebaut de Schotten M. A diffusion tensor imaging tractography atlas for virtual in vivo dissections. Cortex. 2008. 44:1105–1132.51. Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001. 87:198701.52. He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007. 17:2407–2419.53. Kalna G, Higham DJ. Clustering Coefficients for Weighted Networks. Symposium on Network Analysis in Natural Sciences and Engineering. 2006.54. Fransson P, Aden U, Blennow M, Lagercrantz H. The functional architecture of the infant brain as revealed by resting-state FMRI. Cereb Cortex. 2011. 21:145–154.55. Fransson P, Skiold B, Engstrom M, Hallberg B, Mosskin M, Aden U, et al. Spontaneous brain activity in the newborn brain during natural sleep--an fMRI study in infants born at full term. Pediatr Res. 2009. 66:301–305.56. Fransson P, Skiold B, Horsch S, Nordell A, Blennow M, Lagercrantz H, et al. Resting-state networks in the infant brain. Proc Natl Acad Sci U S A. 2007. 104:15531–15536.57. Lin W, Zhu Q, Gao W, Chen Y, Toh CH, Styner M, et al. Functional connectivity MR imaging reveals cortical functional connectivity in the developing brain. AJNR Am J Neuroradiol. 2008. 29:1883–1889.58. Zhang S, Correia S, Laidlaw DH. Identifying white-matter fiber bundles in DTI data using an automated proximity-based fiberclustering method. IEEE Trans Vis Comput Graph. 2008. 14:1044–1053.59. Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Human Brain Mapping. 1999. 7:254–266.60. Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med. 2005. 54:1377–1386.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Effects of Taekwondo Training on Brain Connectivity and Body Intelligence
- Resting-State Electroencephalography (EEG) Functional Connectivity Analysis
- Messages from the Brain Connectivity Regarding Neural Correlates of Consciousness
- Attunement Disorder : A Disorder of Brain Connectivity
- Brain Network Connectivity and Association with Catechol-O-Methyltransferase Gene Polymorphism in Korean Attention-Deficit Hyperactivity Disorder Children