Clin Exp Otorhinolaryngol.
2009 Mar;2(1):44-47. 10.3342/ceo.2009.2.1.44.
Construction of a DNA Chip for Screening of Genetic Hearing Loss
- Affiliations
-
- 1Department of Biology, College of Natural Sciences, Kyungpook National University, Daegu, Korea.
- 2Digital Genomics, Seoul, Korea.
- 3Department of Otorhinolaryngology-Head and Neck Surgery, College of Medicine, Kyungpook National University, Daegu, Korea. leeshu@knu.ac.kr
- KMID: 1466563
- DOI: http://doi.org/10.3342/ceo.2009.2.1.44
Abstract
OBJECTIVES
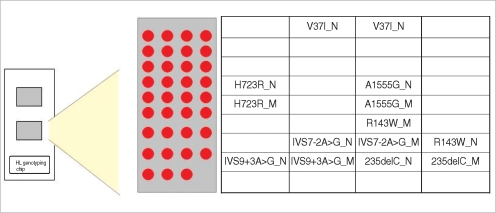
Hearing loss is the most common sensory disorder in humans and genetic causes are estimated to cause more than 50% of all incidents of congenital hearing loss. To develop an efficient method for a genetic diagnosis of hearing loss, we have developed and validated a genetic hearing loss DNA chip that allows the simultaneous analysis of 7 different mutations in the GJB2, SLC26A4, and the mtDNA 12S rRNA genes in Koreans.
METHODS
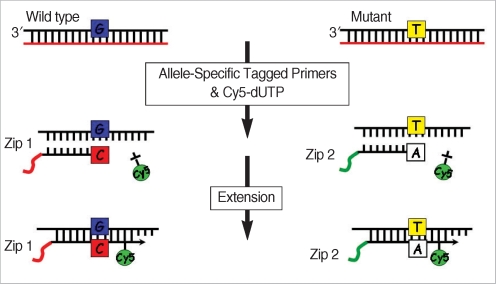
A genotyping microarray, based on the allele-specific primer extension (ASPE) method, was used and preliminary validation was examined from the five patients and five controls that were already known their genotypes by DNA sequencing analysis.
RESULTS
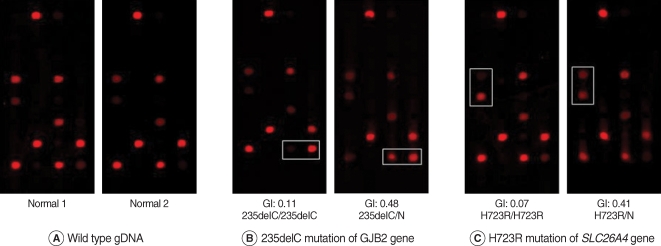
The cutoff Genotyping index (GI) of genotyping for each mutation was set up and validated to discriminate among the genotypes. The result of the DNA chip assay was identical to those of previous results.
CONCLUSION
We successfully designed the genetic hearing loss DNA chip for the first time in Korea and it would be useful for a clinical genetic diagnosis of hearing loss. Further consideration will be needed in order to examine the accuracy of this DNA chip with much larger patient sample numbers.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Genetic Information and Precision Medicine in Hearing Loss
Doo-Yi Oh, Byung Yoon Choi
Clin Exp Otorhinolaryngol. 2020;13(4):315-317. doi: 10.21053/ceo.2020.01606.
Reference
-
1. Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991; 630:16–31. PMID: 1952587.
Article2. Kokotas H, Petersen MB, Willems PJ. Mitochondrial deafness. Clin Genet. 2007; 5. 71(5):379–391. PMID: 17489842.
Article3. Kopp P, Pesce L, Solis-S JC. Pendred syndrome and iodide transport in the thyroid. Trends Endocrinol Metab. 2008; 9. 19(7):260–268. PMID: 18692402.
Article4. Martinez AD, Acuna R, Figueroa V, Maripillan J, Nicholson B. Gap-junction channels dysfunction in deafness and hearing loss. Antioxid Redox Signal. 2009; 2. 11(2):309–322. PMID: 18837651.5. Park HJ, Lee SJ, Jin HS, Lee JO, Go SH, Jang HS, et al. Genetic basis of hearing loss associated with enlarged vestibular aqueducts in Koreans. Clin Genet. 2005; 2. 67(2):160–165. PMID: 15679828.
Article6. Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003; 4. 40(4):242–248. PMID: 12676893.
Article7. Yan D, Park HJ, Ouyang XM, Pandya A, Doi K, Erdenetungalag R, et al. Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in east Asians. Hum Genet. 2003; 12. 114(1):44–50. PMID: 14505035.8. Bae JW, Lee KY, Choi SY, Lee SH, Park HJ, Kim UK. Molecular analysis of mitochondrial gene mutations in Korean patients with nonsyndromic hearing loss. Int J Mol Med. 2008; 8. 22(2):175–180. PMID: 18636170.
Article9. Han SH, Park HJ, Kang EJ, Ryu JS, Lee A, Yang YH, et al. Carrier frequency of GJB2 (connexin-26) mutations causing inherited deafness in the Korean population. J Hum Genet. 2008; 53(11-12):1022–1028. PMID: 19043807.
Article10. Lee KY, Choi SY, Bae JW, Kim S, Chung KW, Drayna D, et al. Molecular analysis of the GJB2, GJB6 and SLC26A4 genes in Korean deafness patients. Int J Pediatr Otorhinolaryngol. 2008; 9. 72(9):1301–1309. PMID: 18585793.
Article11. Park HJ, Hahn SH, Chun YM, Park K, Kim HN. Connexin26 mutations associated with nonsyndromic hearing loss. Laryngoscope. 2000; 9. 110(9):1535–1538. PMID: 10983956.
Article12. Kelley PM, Harris DJ, Comer BC, Askew JW, Fowler T, Smith SD, et al. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am J Hum Genet. 1998; 4. 62(4):792–799. PMID: 9529365.
Article13. Snoeckx RL, Huygen PL, Feldmann D, Marlin S, Denoyelle F, Waligora J, et al. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet. 2005; 12. 77(6):945–957. PMID: 16380907.14. Brobby GW, Müller-Myhsok B, Horstmann RD. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N Engl J Med. 1998; 2. 19. 338(8):548–550. PMID: 9471561.
Article15. Dai P, Yu F, Han B, Wu H, Yuan YY, Li Q, et al. Features of nationwide distribution and frequency of a common gap junction beta-2 gene mutation in China. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2007; 11. 42(11):804–808. PMID: 18300439.16. Coucke PJ, Van Hauwe P, Everett LA, Demirhan O, Kabakkaya Y, Dietrich NL, et al. Identification of two different mutations in the PDS gene in an inbred family with Pendred syndrome. J Med Genet. 1999; 6. 36(6):475–477. PMID: 10874637.17. Usami S, Abe S, Weston MD, Shinkawa H, Van Camp G, Kimberling WJ. Non-syndromic hearing loss associated with enlarged vestibular aqueduct is caused by PDS mutations. Hum Genet. 1999; 2. 104(2):188–192. PMID: 10190331.
Article18. Siemering K, Manji SS, Hutchison WM, Du Sart D, Phelan D, Dahl HH. Detection of mutations in genes associated with hearing loss using a microarray-based approach. J Mol Diagn. 2006; 9. 8(4):483–489. PMID: 16931589.
Article19. Usami S, Abe S, Akita J, Namba A, Shinkawa H, Ishii M, et al. Prevalence of mitochondrial gene mutations among hearing impaired patients. J Med Genet. 2000; 1. 37(1):38–40. PMID: 10633132.
Article20. Cremers FP, Kimberling WJ, Kulm M, de Brouwer AP, van Wijk E, te Brinke H, et al. Development of a genotyping microarray for Usher syndrome. J Med Genet. 2007; 2. 44(2):153–160. PMID: 16963483.
Article21. Gardner P, Oitmaa E, Messner A, Hoefsloot L, Metspalu A, Schrijver I. Simultaneous multigene mutation detection in patients with sensorineural hearing loss through a novel diagnostic microarray: a new approach for newborn screening follow-up. Pediatrics. 2006; 9. 118(3):985–994. PMID: 16950989.
Article