Healthc Inform Res.
2013 Sep;19(3):177-185. 10.4258/hir.2013.19.3.177.
Real-Data Comparison of Data Mining Methods in Prediction of Diabetes in Iran
- Affiliations
-
- 1Department of Biostatistics, School of Public Health, Hamadan University of Medical Sciences, Hamadan, Iran.
- 2Research Center for Health Sciences and Department of Epidemiology & Biostatistics, School of Public Health, Hamadan University of Medical Sciences, Hamadan, Iran. mahjub@umsha.ac.ir
- 3Department of Science, Hamadan University of Technology, Hamadan, Iran.
- KMID: 2166696
- DOI: http://doi.org/10.4258/hir.2013.19.3.177
Abstract
OBJECTIVES
Diabetes is one of the most common non-communicable diseases in developing countries. Early screening and diagnosis play an important role in effective prevention strategies. This study compared two traditional classification methods (logistic regression and Fisher linear discriminant analysis) and four machine-learning classifiers (neural networks, support vector machines, fuzzy c-mean, and random forests) to classify persons with and without diabetes.
METHODS
The data set used in this study included 6,500 subjects from the Iranian national non-communicable diseases risk factors surveillance obtained through a cross-sectional survey. The obtained sample was based on cluster sampling of the Iran population which was conducted in 2005-2009 to assess the prevalence of major non-communicable disease risk factors. Ten risk factors that are commonly associated with diabetes were selected to compare the performance of six classifiers in terms of sensitivity, specificity, total accuracy, and area under the receiver operating characteristic (ROC) curve criteria.
RESULTS
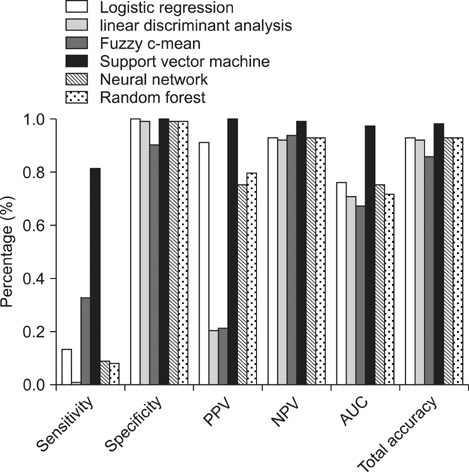
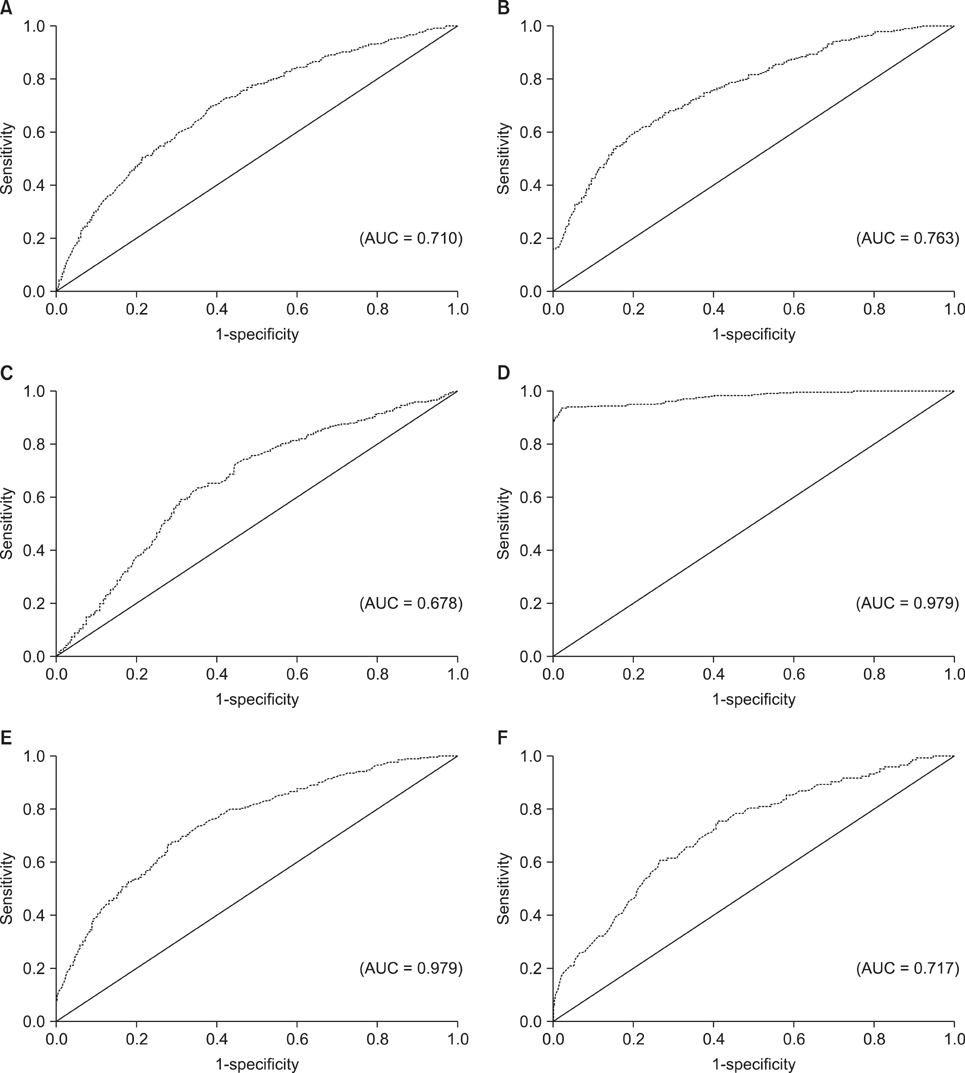
Support vector machines showed the highest total accuracy (0.986) as well as area under the ROC (0.979). Also, this method showed high specificity (1.000) and sensitivity (0.820). All other methods produced total accuracy of more than 85%, but for all methods, the sensitivity values were very low (less than 0.350).
CONCLUSIONS
The results of this study indicate that, in terms of sensitivity, specificity, and overall classification accuracy, the support vector machine model ranks first among all the classifiers tested in the prediction of diabetes. Therefore, this approach is a promising classifier for predicting diabetes, and it should be further investigated for the prediction of other diseases.
MeSH Terms
Figure
Cited by 2 articles
-
Prediction of Serum Creatinine in Hemodialysis Patients Using a Kernel Approach for Longitudinal Data
Mohammad Moqaddasi Amiri, Leili Tapak, Javad Faradmal, Javad Hosseini, Ghodratollah Roshanaei
Healthc Inform Res. 2020;26(2):112-118. doi: 10.4258/hir.2020.26.2.112.Prediction of Kidney Graft Rejection Using Artificial Neural Network
Leili Tapak, Omid Hamidi, Payam Amini, Jalal Poorolajal
Healthc Inform Res. 2017;23(4):277-284. doi: 10.4258/hir.2017.23.4.277.
Reference
-
1. International Diabetes Federation. IDF Diabetes Atlas: the global burden [Internet]. Brussels, Belgium: International Diabetes Federation;c2013. cited at 2013 Sep 1. Available from: http://www.idf.org/diabetesatlas/5e/the-global-burden.2. Priya R, Aruna P. Review of automated diagnosis of diabetic retinopathy using the support vector machine. Int J Appl Eng Res (Dindigul). 2011; 1(4):844–863.3. Calder R, Alexander C. Cardiovascular disease in people with diabetes mellitus. Pract Diabetol. 2000; 19(4):7–18.4. Barr EL, Zimmet PZ, Welborn TA, Jolley D, Magliano DJ, Dunstan DW, et al. Risk of cardiovascular and all-cause mortality in individuals with diabetes mellitus, impaired fasting glucose, and impaired glucose tolerance: the Australian Diabetes, Obesity, and Lifestyle Study (AusDiab). Circulation. 2007; 116(2):151–157.
Article5. Pi-Sunyer FX. How effective are lifestyle changes in the prevention of type 2 diabetes mellitus? Nutr Rev. 2007; 65(3):101–110.
Article6. IDF Clinical Guidelines Task Force. Global Guideline for Type 2 Diabetes: recommendations for standard, comprehensive, and minimal care. Diabet Med. 2006; 23(6):579–593.7. Thomas C, Hypponen E, Power C. Type 2 diabetes mellitus in midlife estimated from the Cambridge Risk Score and body mass index. Arch Intern Med. 2006; 166(6):682–688.
Article8. Maroco J, Silva D, Rodrigues A, Guerreiro M, Santana I, de Mendonca A. Data mining methods in the prediction of Dementia: a real-data comparison of the accuracy, sensitivity and specificity of linear discriminant analysis, logistic regression, neural networks, support vector machines, classification trees and random forests. BMC Res Notes. 2011; 4:299.
Article9. Yu W, Liu T, Valdez R, Gwinn M, Khoury MJ. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and prediabetes. BMC Med Inform Decis Mak. 2010; 10:16.
Article10. Kim S, Kim W, Park RW. A comparison of intensive care unit mortality prediction models through the use of data mining techniques. Healthc Inform Res. 2011; 17(4):232–243.
Article11. Son YJ, Kim HG, Kim EH, Choi S, Lee SK. Application of support vector machine for prediction of medication adherence in heart failure patients. Healthc Inform Res. 2010; 16(4):253–259.
Article12. Lee SK, Kang BY, Kim HG, Son YJ. Predictors of medication adherence in elderly patients with chronic diseases using support vector machine models. Healthc Inform Res. 2013; 19(1):33–41.
Article13. Lehmann C, Koenig T, Jelic V, Prichep L, John RE, Wahlund LO, et al. Application and comparison of classification algorithms for recognition of Alzheimer's disease in electrical brain activity (EEG). J Neurosci Methods. 2007; 161(2):342–350.
Article14. Hachesu PR, Ahmadi M, Alizadeh S, Sadoughi F. Use of data mining techniques to determine and predict length of stay of cardiac patients. Healthc Inform Res. 2013; 19(2):121–129.
Article15. Priya R, Aruna P. SVM and neural network based diagnosis of diabetic retinopathy. Int J Comput Appl. 2012; 41(1):6–12.
Article16. Finch H, Schneider MK. Classification accuracy of neural networks vs. discriminant analysis, logistic regression, and classification and regression trees: three- and five-group cases. Methodology (Gott). 2007; 3(2):47–57.
Article17. Austin PC. A comparison of regression trees, logistic regression, generalized additive models, and multivariate adaptive regression splines for predicting AMI mortality. Stat Med. 2007; 26(15):2937–2957.
Article18. Gelnarova E, Safarik L. Comparison of three statistical classifiers on a prostate cancer data. Neural Netw World. 2005; 15(4):311–318.19. Green M, Bjork J, Forberg J, Ekelund U, Edenbrandt L, Ohlsson M. Comparison between neural networks and multiple logistic regression to predict acute coronary syndrome in the emergency room. Artif Intell Med. 2006; 38(3):305–318.
Article20. Meyer D, Leisch F, Hornik K. The support vector machine under test. Neurocomputing. 2003; 55(1-2):169–186.
Article21. Poorolajal J, Zamani R, Mir-Moeini R, Amiri B, Majzoobi M, Erfani H, et al. Five-year evaluation of chronic diseases in Hamadan, Iran: 2005-2009. Iran J Public Health. 2012; 41(3):71–81.22. Little RJ. A test of missing completely at random for multivariate data with missing values. J Am Stat Assoc. 1988; 83(404):1198–1202.
Article23. Yoon H, Jun SC, Hyun Y, Bae GO, Lee KK. A comparative study of artificial neural networks and support vector machines for predicting groundwater levels in a coastal aquifer. J Hydrol. 2011; 396(1-2):128–138.
Article24. The Comprehensive R Archive Network (CRAN) package [Internet]. The R Foundation;cited at 2013 Sep 1. Available from: http://cran.r-project.org/web/packages/.25. Schwarzer G, Vach W, Schumacher M. On the misuses of artificial neural networks for prognostic and diagnostic classification in oncology. Stat Med. 2000; 19(4):541–561.
Article26. Oliveira PP Jr, Nitrini R, Busatto G, Buchpiguel C, Sato JR, Amaro E Jr. Use of SVM methods with surface-based cortical and volumetric subcortical measurements to detect Alzheimer's disease. J Alzheimers Dis. 2010; 19(4):1263–1272.
Article27. Zhu Y, Tan Y, Hua Y, Wang M, Zhang G, Zhang J. Feature selection and performance evaluation of support vector machine (SVM)-based classifier for differentiating benign and malignant pulmonary nodules by computed tomography. J Digit Imaging. 2010; 23(1):51–65.
Article28. Abbasimehr H, Setak M, Tarokh MJ. A neuro-fuzzy classifier for customer churn prediction. Int J Comput Appl. 2011; 19(8):35–41.29. Breiman L. Random forests. Mach Learn. 2001; 45(1):5–32.30. Smith A, Sterba-Boatwright B, Mott J. Novel application of a statistical technique, random forests, in a bacterial source tracking study. Water Res. 2010; 44(14):4067–4076.
Article31. Auria L, Moro RA. Support vector machines (SVM) as a technique for solvency analysis. Berlin, Germany: Deutsches Institut fur Wirtschaftsforschung;2008.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Real-Data Comparison of Data Mining Methods in Prediction of Diabetes in Iran
- Data Mining for High Dimensional Data in Drug Discovery and Development
- Prediction of Diabetes Using Data Mining and Machine Learning Algorithms: A Cross-Sectional Study
- Systematic Review of Data Mining Applications in Patient-Centered Mobile-Based Information Systems
- The Efficient Ways of Health Promotion Program on Public Health Center Using Data Mining