Healthc Inform Res.
2013 Jun;19(2):102-109. 10.4258/hir.2013.19.2.102.
Lessons Learned from Development of De-identification System for Biomedical Research in a Korean Tertiary Hospital
- Affiliations
-
- 1Department of Biomedical Informatics, Asan Medical Center, Seoul, Korea.
- 2Office of Clinical Research Information, Asan Medical Center, Seoul, Korea. jaeholee@amc.seoul.kr
- 3University of Ulsan College of Medicine, Seoul, Korea.
- 4Department of Pulmonary & Critical Care Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- 5Department of Emergency Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 2229491
- DOI: http://doi.org/10.4258/hir.2013.19.2.102
Abstract
OBJECTIVES
The Korean government has enacted two laws, namely, the Personal Information Protection Act and the Bioethics and Safety Act to prevent the unauthorized use of medical information. To protect patients' privacy by complying with governmental regulations and improve the convenience of research, Asan Medical Center has been developing a de-identification system for biomedical research.
METHODS
We reviewed Korean regulations to define the scope of the de-identification methods and well-known previous biomedical research platforms to extract the functionalities of the systems. Based on these review results, we implemented necessary programs based on the Asan Medical Center Information System framework which was built using the Microsoft. NET Framework and C#.
RESULTS
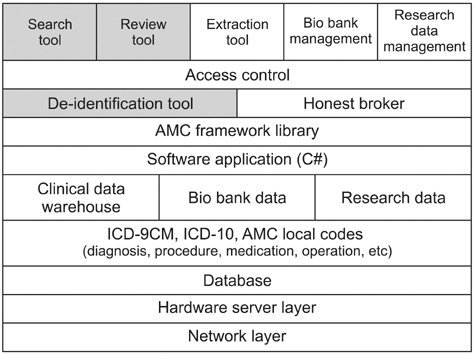
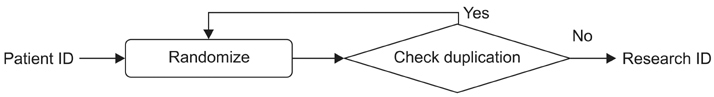
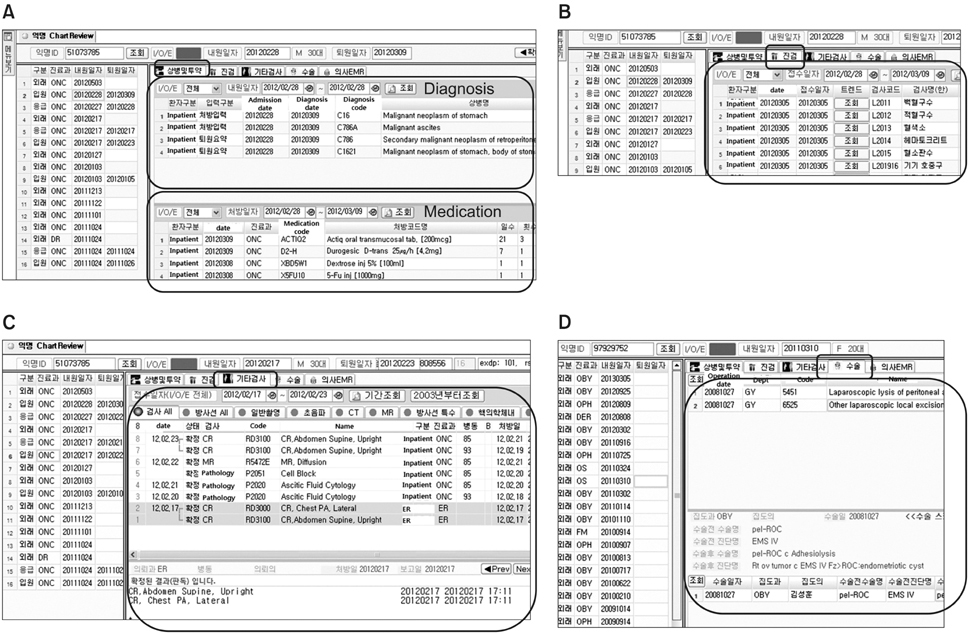
The developed de-identification system comprises three main components: a de-identification tool, a search tool, and a chart review tool. The de-identification tool can substitute a randomly assigned research ID for a hospital patient ID, remove the identifiers in the structured format, and mask them in the unstructured format, i.e., texts. This tool achieved 98.14% precision and 97.39% recall for 6,520 clinical notes. The search tool can find the number of patients which satisfies given search criteria. The chart review tool can provide de-identified patient's clinical data for review purposes.
CONCLUSIONS
We found that a clinical data warehouse was essential for successful implementation of the de-identification system, and this system should be tightly linked to an electronic Institutional Review Board system for easy operation of honest brokers. Additionally, we found that a secure cloud environment could be adopted to protect patients' privacy more thoroughly.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Dementia Research Using Healthcare Big Data
Hun-Sung Kim, Dai-Jin Kim
Dement Neurocogn Disord. 2019;18(3):73-76. doi: 10.12779/dnd.2019.18.3.73.
Reference
-
1. Office for Civil Rights. Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule [Internet]. Washington (DC): US Department of Health & Human Service;c2013. cited at 2013 Mar 28. Available from: http://www.hhs.gov/ocr/privacy/hipaa/understanding/coveredentities/De-identification/hhs_deid_guidance.pdf.2. Korea Ministry of Government Legislation. The Personal Information Protection Act [Internet]. Seoul, Korea: Korea Ministry of Government Legislation;c2013. cited at 2013 Mar 28. Available from: http://www.law.go.kr/lsEfInfoP.do?lsiSeq=136728#0000.3. Korea Ministry of Government Legislation. The Bioethics and Safety Act [Internet]. Seoul, Korea: Korea Ministry of Government Legislation;c2013. cited at 2013 Mar 28. Available from: http://www.law.go.kr/lsEfInfoP.do?lsiSeq=137037#0000.4. Ryu HJ, Kim WS, Lee JH, Min SW, Kim SJ, Lee YS, et al. Asan medical information system for healthcare quality improvement. Healthc Inform Res. 2010; 16(3):191–197.
Article5. Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE: an integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009; 2009:391–395.6. Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc. 2010; 17(2):124–130.
Article7. Nalichowski R, Keogh D, Chueh HC, Murphy SN. Calculating the benefits of a Research Patient Data Repository. AMIA Annu Symp Proc. 2006; 1044.8. Stanford Center for Clinical Informatics, Stanford School of Medicine. Stanford Translational Research Integrated Database Environment (STRIDE) [Internet]. Stanford (CA): Stanford School of Medicine;c2013. cited at 2013 Mar 28. Available from: https://clinicalinformatics.stanford.edu/research/stride.html.9. National Center for Biomedical Computing. Informatics for Integrating Biology & the Bedside [Internet]. Bethesda (MD): National Institutes of Health;c2013. cited at 2013 Mar 28. Available from: https://www.i2b2.org/.10. Partners Healthcare. Research Patient Data Registry (RPDR) [Internet]. Boston (MA): Partners HealthCare;c2013. cited at 2013 March 28. Available from: http://rc.partners.org/rpdr.11. El Emam K. Methods for the de-identification of electronic health records for genomic research. Genome Med. 2011; 3(4):25.
Article12. Fraser R, Willison D. Tools for de-identification of personal health information. [Toronto]: Pan Canadian Health Information Privacy Group;2009. cited at 2013 Apr 3. Available from: https://www.infoway-inforoute.ca/index.php/resources/video-gallery/doc_download/624-tools-for-de-identification-of-pseronal-health-information.13. Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. 2010; 10:70.
Article14. Deleger L, Molnar K, Savova G, Xia F, Lingren T, Li Q, et al. Large-scale evaluation of automated clinical note de-identification and its impact on information extraction. J Am Med Inform Assoc. 2013; 20(1):84–94.
Article15. Shin SY, Shin Y, Choi HJ, Park J, Lyu LM, Kim WS, et al. Deidentification method for bilingual EMR free texts. In : Proceedings of the 14th World Congress on Medical and Health Informatics; 2013 Aug 20-23; Copenhagen, Denmark.16. El Emam K. Heuristics for de-identifying health data. IEEE Secur Priv. 2008; 6(4):58–61.
Article17. El Emam K, Dankar FK, Issa R, Jonker E, Amyot D, Cogo E, et al. A globally optimal k-anonymity method for the de-identification of health data. J Am Med Inform Assoc. 2009; 16(5):670–682.
Article18. Liu J, Erdal S, Silvey SA, Ding J, Riedel JD, Marsh CB, et al. Toward a fully de-identified biomedical information warehouse. AMIA Annu Symp Proc. 2009; 2009:370–374.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correction: Lessons Learned from Development of De-identification System for Biomedical Research in a Korean Tertiary Hospital
- Lessons Learned from Development of De-identification System for Biomedical Research in a Korean Tertiary Hospital
- Top Ten Lessons Learned from Trials in Oligometastatic Cancers
- Lessons learned with a longer follow-up from SOLO 1
- Implementation and lessons learned from 2 online interprofessional faculty development programs for improving educational practice in the health professions in Chile and the United Kingdom from 2018 to 2021