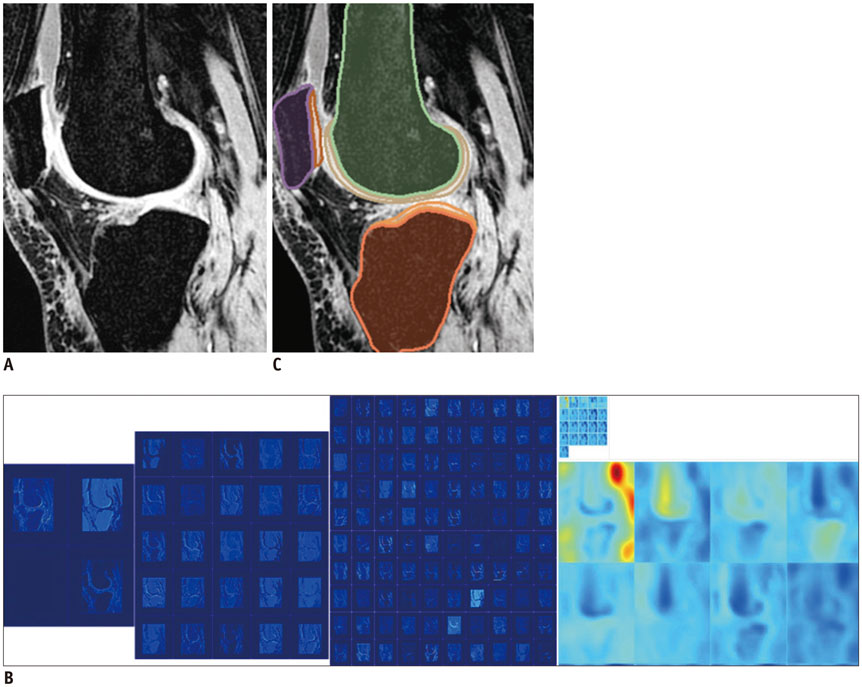
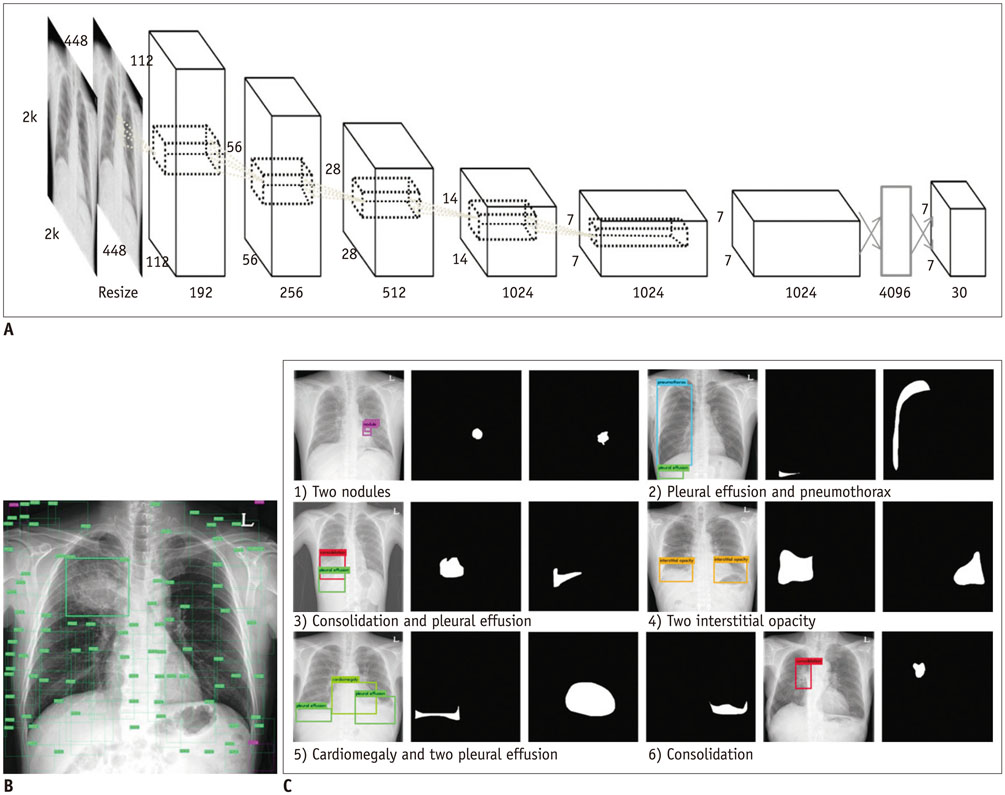
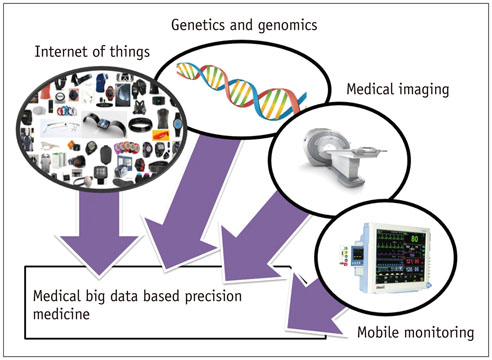
Deep Learning in Medical Imaging: General Overview
- Affiliations
-
- 1Biomedical Engineering Research Center, University of Ulsan College of Medicine, Asan Medical Center, Seoul 05505, Korea.
- 2Department of Radiology, Research Institute of Radiology, University of Ulsan College of Medicine, Asan Medical Center, Seoul 05505, Korea. namkugkim@gmail.com
- 3Department of Convergence Medicine, Biomedical Engineering Research Center, University of Ulsan College of Medicine, Asan Medical Center, Seoul 05505, Korea. joonbeomseo@gmail.com
- KMID: 2427226
- DOI: http://doi.org/10.3348/kjr.2017.18.4.570
Abstract
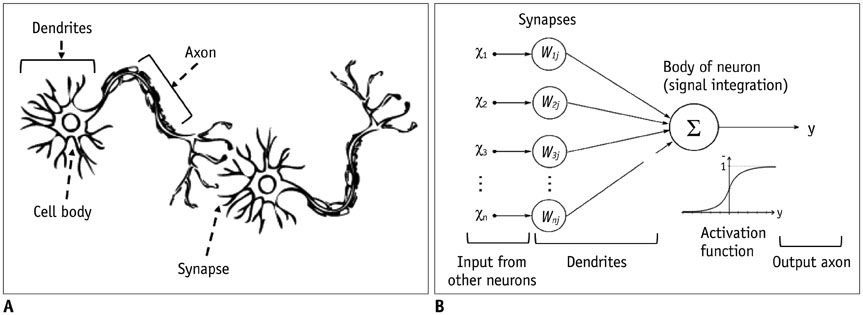
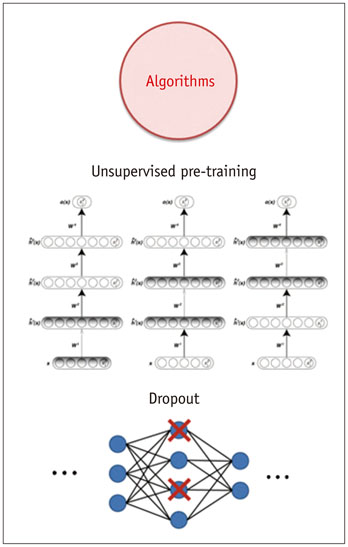
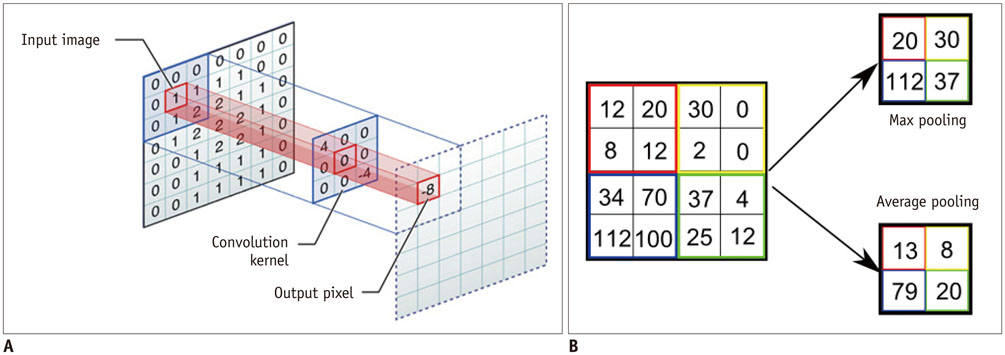
- The artificial neural network (ANN)-a machine learning technique inspired by the human neuronal synapse system-was introduced in the 1950s. However, the ANN was previously limited in its ability to solve actual problems, due to the vanishing gradient and overfitting problems with training of deep architecture, lack of computing power, and primarily the absence of sufficient data to train the computer system. Interest in this concept has lately resurfaced, due to the availability of big data, enhanced computing power with the current graphics processing units, and novel algorithms to train the deep neural network. Recent studies on this technology suggest its potentially to perform better than humans in some visual and auditory recognition tasks, which may portend its applications in medicine and healthcare, especially in medical imaging, in the foreseeable future. This review article offers perspectives on the history, development, and applications of deep learning technology, particularly regarding its applications in medical imaging.
Keyword
MeSH Terms
Figure
Cited by 16 articles
-
Connecting Technological Innovation in Artificial Intelligence to Real-world Medical Practice through Rigorous Clinical Validation: What Peer-reviewed Medical Journals Could Do
Seong Ho Park, Herbert Y. Kressel
J Korean Med Sci. 2018;33(22):. doi: 10.3346/jkms.2018.33.e152.Feasibility of fully automated classification of whole slide images based on deep learning
Kyung-Ok Cho, Sung Hak Lee, Hyun-Jong Jang
Korean J Physiol Pharmacol. 2020;24(1):89-99. doi: 10.4196/kjpp.2020.24.1.89.Basics of Deep Learning: A Radiologist's Guide to Understanding Published Radiology Articles on Deep Learning
Synho Do, Kyoung Doo Song, Joo Won Chung
Korean J Radiol. 2020;21(1):33-41. doi: 10.3348/kjr.2019.0312.The Role of medical doctor in the era of artificial intelligence
Joon Beom Seo
J Korean Med Assoc. 2019;62(3):136-139. doi: 10.5124/jkma.2019.62.3.136.Age of Data in Contemporary Research Articles Published in Representative General Radiology Journals
Ji Hun Kang, Dong Hwan Kim, Seong Ho Park, Jung Hwan Baek
Korean J Radiol. 2018;19(6):1172-1178. doi: 10.3348/kjr.2018.19.6.1172.Fully Automatic Segmentation of Acute Ischemic Lesions on Diffusion-Weighted Imaging Using Convolutional Neural Networks: Comparison with Conventional Algorithms
Ilsang Woo, Areum Lee, Seung Chai Jung, Hyunna Lee, Namkug Kim, Se Jin Cho, Donghyun Kim, Jungbin Lee, Leonard Sunwoo, Dong-Wha Kang
Korean J Radiol. 2019;20(8):1275-1284. doi: 10.3348/kjr.2018.0615.CT Image Conversion among Different Reconstruction Kernels without a Sinogram by Using a Convolutional Neural Network
Sang Min Lee, June-Goo Lee, Gaeun Lee, Jooae Choe, Kyung-Hyun Do, Namkug Kim, Joon Beom Seo
Korean J Radiol. 2019;20(2):295-303. doi: 10.3348/kjr.2018.0249.Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers
Dong Wook Kim, Hye Young Jang, Kyung Won Kim, Youngbin Shin, Seong Ho Park
Korean J Radiol. 2019;20(3):405-410. doi: 10.3348/kjr.2019.0025.Effect of a Deep Learning Framework-Based Computer-Aided Diagnosis System on the Diagnostic Performance of Radiologists in Differentiating between Malignant and Benign Masses on Breast Ultrasonography
Ji Soo Choi, Boo-Kyung Han, Eun Sook Ko, Jung Min Bae, Eun Young Ko, So Hee Song, Mi-ri Kwon, Jung Hee Shin, Soo Yeon Hahn
Korean J Radiol. 2019;20(5):749-758. doi: 10.3348/kjr.2018.0530.Radiomics and Deep Learning: Hepatic Applications
Hyo Jung Park, Bumwoo Park, Seung Soo Lee
Korean J Radiol. 2020;21(4):387-401. doi: 10.3348/kjr.2019.0752.Principles for evaluating the clinical implementation of novel digital healthcare devices
Seong Ho Park, Kyung-Hyun Do, Joon-Il Choi, Jung Suk Sim, Dal Mo Yang, Hong Eo, Hyunsik Woo, Jeong Min Lee, Seung Eun Jung, Joo Hyeong Oh
J Korean Med Assoc. 2018;61(12):765-775. doi: 10.5124/jkma.2018.61.12.765.Artificial Intelligence in Medicine: Beginner's Guide
Seong Ho Park
J Korean Soc Radiol. 2018;78(5):301-308. doi: 10.3348/jksr.2018.78.5.301.Imaging Informatics: A New Horizon for Radiologyin the Era of Artificial Intelligence, Big Data, and Data Science
Jong Hyo Kim
J Korean Soc Radiol. 2019;80(2):176-201. doi: 10.3348/jksr.2019.80.2.176.Application of Artificial Screening Intelligence in Lung Cancer
Sang Min Lee, Chang Min Park
J Korean Soc Radiol. 2019;80(5):872-879. doi: 10.3348/jksr.2019.80.5.872.What should medical students know about artificial intelligence in medicine?
Seong Ho Park, Kyung-Hyun Do, Sungwon Kim, Joo Hyun Park, Young-Suk Lim, A Ra Cho
J Educ Eval Health Prof. 2019;16:18. doi: 10.3352/jeehp.2019.16.18.Convolutional Neural Network Technology in Endoscopic Imaging: Artificial Intelligence for Endoscopy
Joonmyeong Choi, Keewon Shin, Jinhoon Jung, Hyun-Jin Bae, Do Hoon Kim, Jeong-Sik Byeon, Namku Kim
Clin Endosc. 2020;53(2):117-126. doi: 10.5946/ce.2020.054.
Reference
-
1. Murphy KP. Machine learning: a probabilistic perspective. 1st ed. Cambridge: The MIT Press;2012. p. 25.2. Fenton JJ, Taplin SH, Carney PA, Abraham L, Sickles EA, D'Orsi C, et al. Influence of computer-aided detection on performance of screening mammography. N Engl J Med. 2007; 356:1399–1409.3. Lehman CD, Wellman RD, Buist DS, Kerlikowske K, Tosteson AN, Miglioretti DL, et al. Diagnostic accuracy of digital screening mammography with and without computer-aided detection. JAMA Intern Med. 2015; 175:1828–1837.4. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007; 73:5261–5267.5. Byvatov E, Fechner U, Sadowski J, Schneider G. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J Chem Inf Comput Sci. 2003; 43:1882–1889.6. In : Tong S, Chang E, editors. Support vector machine active learning for image retrieval. Proceedings of the 9th ACM International Conference on Multimedia; 2001 October 5-September 30; Ottawa, Canada. New York: ACM;2001. p. 107.7. Arbib MA. The handbook of brain theory and neural networks. 2nd ed. Boston: The MIT Press;2003.8. Drucker H, Burges CJC, Kaufman L, Smola A, Vapnik V. Support vector regression machines. Adv Neural Inf Process Syst. 1997; 9:155–161.9. Yu PS, Chen ST, Chang IF. Support vector regression for real-time flood stage forecasting. J Hydrol (Amst). 2006; 328:704–716.10. Tay FEH, Cao L. Application of support vector machines in financial time series forecasting. Omega (Westport). 2001; 29:309–317.11. Haykin SS. Neural networks: a comprehensive foundation. New York: Macmillan College Publishing;1994. p. 107–116.12. Kiwiel KC. Convergence and efficiency of subgradient methods for quasiconvex minimization. Math Program. 2001; 90:1–25.13. In : Deng L, Hinton G, Kingsbury B, editors. New types of deep neural network learning for speech recognition and related applications: an overview. Proceedings of 2013 IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP); 2013 May 26-31; Vancouver, Canada. IEEE;2013. p. 8599–8603.14. Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006; 18:1527–1554.15. Playground.tensorflow.org Web site. A Neural Network Playground-TensorFlow. Accessed April 1, 2017. http://playground.tensorflow.org.16. In : Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, editors. Caffe: convolutional architecture for fast feature embedding. Proceedings of the 22nd ACM International Conference on Multimedia; 2014 November 3-7; Orlando, FL, USA. New York: ACM;2014. p. 675–678.17. Yu D, Eversole A, Seltzer ML, Yao K, Huang Z, Guenter B, et al. An introduction to computational networks and the computational network toolkit. New York: Microsoft Research;2014.18. In : Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, editors. TensorFlow: a system for large-scale machine learning. Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation; 2016 November 2-4; Savannah, GA, USA. Berkeley: USENIX Association;2016. p. 265–283.19. The Theano Development Team. Al-Rfou R, Alain G, Almahairi A, Angermueller C, Bahdanau D, et al. Theano: a python framework for fast computation of mathematical expressions. ArXiv org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1605.02688.20. In : Collobert R, van der Maaten L, Joulin A, editors. Torchnet: an open-source platform for (deep) learning research. Proceedings of the 33rd International Conference on Machine Learning (ICML-2016); 2016 June 19-24; New York, NY, USA. New York: JMLR;2016.21. Hubel DH, Wiesel TN. Receptive fields and functional architecture of monkey striate cortex. J Physiol. 1968; 195:215–243.22. Ujjwalkarn.me Web site. An intuitive explanation of convolutional neural networks. Accessed April 1, 2017. https://ujjwalkarn.me/2016/08/11/intuitive-explanation-convnets.23. In : Krizhevsky A, Sutskever I, Hinton GE, editors. ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems; 2012 December 3-6; Lake Tahoe, NV, USA. Curran Associates Inc.;2012. p. 1–9.24. In : Mikolov T, Karafiát M, Burget L, Cernocký J, Khudanpur S, editors. Recurrent neural network based language model. Proceedings of the 11th Annual Conference of the International Speech Communication Association (INTERSPEECH 2010); 2010 September 26-30; Makuhari, Japan. International Speech Communication Association;2010. p. 1045–1048.25. In : Gregor K, Danihelka I, Graves A, Rezende DJ, Wierstra D, editors. DRAW: a recurrent neural network for image generation. Proceedings of the 32nd International Conference on Machine Learning (ICML-2015); 2015 July 6-11; Lille, France. JMLR;2015. p. 1462–1471.26. Cireşan D, Meier U, Masci J, Schmidhuber J. Multi-column deep neural network for traffic sign classification. Neural Netw. 2012; 32:333–338.27. Ning F, Delhomme D, LeCun Y, Piano F, Bottou L, Barbano PE. Toward automatic phenotyping of developing embryos from videos. IEEE Trans Image Process. 2005; 14:1360–1371.28. Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Semantic image segmentation with deep convolutional nets and fully connected CRFs. ArXiv.org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1412.7062.29. Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015; 115:211–252.30. Dahl GE, Sainath TN, Hinton GE. Improving deep neural networks for LVCSR using rectified linear units and dropout. In : Proceedings of the 38th IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP '13); 2013 May 26-31; Vancouver, Canada. IEEE;2013. p. 8609–8613.31. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In : Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in neural information processing system 25. Nevada: Curran Associates Inc.;2012. p. 1097–1105.32. Image-net.org Website. Large Scale Visual Recognition Challenge 2016 (ILSVRC2016) results. Accessed April 1, 2017. http://image-net.org/challenges/LSVRC/2016/.33. In : Ren S, He K, Girshick R, Sun J, editors. Faster R-CNN: towards real-time object detection with region proposal networks. Proceedings of the 28th International Conference on Neural Information Processing Systems (NIPS-2015); 2015 December 7-12; Montreal, Canada. Cambridge: MIT Press;2015. p. 91–99.34. Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017; 39:640–651.35. In : Johnson J, Karpathy A, Fei LF, editors. DenseCap: fully convolutional localization networks for dense captioning. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 June 27-30; Seattle, WA, USA. IEEE;2016. p. 4565–4574.36. In : Antol S, Agrawal A, Lu J, Mitchell M, Batra D, Zitnick CL, editors. VQA: visual question answering. Proceedings of the IEEE International Conference on Computer Vision (ICCV); 2015 December 7-13; Santiago, Chile. IEEE;2015. p. 2425–2433.37. Antoniol G, Fiutem R, Flor R, Lazzari G. Radiological reporting based on voice recognition. In : In : Bass LJ, Gornostaev J, Unger C, editors. Human-Computer Interaction. Proceedings of the third International Conference, EWHCI '93 Moscow; 1993 August 3–7; Moscow, Russia. Moscow: Springer;1993. p. 242–253.38. Antoniol G, Brugnara F, Dalla Palma F, Lazzari G, Moser E. A.RE.S.: an interface for automatic reporting by speech. In : Adlassnig KP, Grabner G, Bengtsson S, Hansen R, editors. Proceedings of Medical Informatics Europe 1991. 1991 August 19-22; Vienna, Austria. Berlin, Heidelberg: Springer-Verlag;1991. p. 150–154.39. Bahl LR, Jelinek F, Mercer RL. A maximum likelihood approach to continuous speech recognition. IEEE Trans Pattern Anal Mach Intell. 1983; 5:179–190.40. Baker JK. Trainable grammars for speech recognition. J Acoust Soc Am. 1979; 65:S1. S132.41. Middleton I, Damper RI. Segmentation of magnetic resonance images using a combination of neural networks and active contour models. Med Eng Phys. 2004; 26:71–86.42. Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016; 35:1240–1251.43. Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJ, Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016; 35:1252–1261.44. In : Ciresan D, Giusti A, Gambardella LM, Schmidhuber J, editors. Deep neural networks segment neuronal membranes in electron microscopy images. Proceedings of the 25th International Conference on Neural Information Processing Systems; 2012 December 3-6; Lake Tahoe, NV, USA. Curran Associates Inc.;2012. p. 2843–2851.45. Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv. 2013; 16(Pt 2):246–253.46. Glavan CC, Holban S. Segmentation of bone structure in X-ray images using convolutional neural network. Adv Electr Comput Eng. 2013; 13:87–94.47. Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv. 2013; 16(Pt 2):411–418.48. Kang K, Wang X. Fully convolutional neural networks for crowd segmentation. ArXiv.org Web site. Accessed April 1, 2017. http://arxiv.org/abs/1411.4464.49. Roura E, Oliver A, Cabezas M, Valverde S, Pareto D, Vilanova JC, et al. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology. 2015; 57:1031–1043.50. Brosch T, Yoo Y, Tang LYW, Li DKB, Traboulsee A, Tam R. Deep convolutional encoder networks for multiple sclerosis lesion segmentation. In : Navab N, Hornegger J, Wells WM, Frangi AF, editors. Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015. New York. Spinger;2015. p. 3–11.51. Brosch T, Tang LY, Yoo Y, Li DK, Traboulsee A, Tam R. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging. 2016; 35:1229–1239.52. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998; 86:2278–2324.53. In : Zeiler MD, Taylor GW, Fergus R, editors. Adaptive deconvolutional networks for mid and high level feature learning. Proceedings of the 2011 International Conference on Computer Vision; 2011 November 6-13; Barcelona, Spain. Washington, DC: IEEE Computer Society;2011. p. 2018–2025.54. In : Wohlhart P, Lepetit V, editors. Learning descriptors for object recognition and 3D pose estimation. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 3109–3118.55. In : Dollár P, Welinder P, Perona P, editors. Cascaded pose regression. Proceedings of 2010 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2010 June 13-18; San Francisco, CA, USA. IEEE;2010. p. 1078–1085.56. In : Zach C, Sanchez AP, Pham MT, editors. A dynamic programming approach for fast and robust object pose recognition from range images. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 196–203.57. In : Mottaghi R, Xiang Y, Savarese S, editors. A coarse-to-fine model for 3D pose estimation and sub-category recognition. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 418–426.58. Shun Miao, Wang ZJ, Rui Liao. A CNN regression approach for real-time 2D/3D registration. IEEE Trans Med Imaging. 2016; 35:1352–1363.59. Zhao L, Jia K. Deep Adaptive Log-demons: diffeomorphic image registration with very large deformations. Comput Math Methods Med. 2015; 2015:836202.60. Mao J, Xu W, Yang Y, Wang J, Huang Z, Yuille A. Explain images with multimodal recurrent neural networks. ArXiv.org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1410.1090.61. Socher R, Karpathy A, Le QV, Manning CD, Ng AY. Grounded compositional semantics for finding and describing images with sentences. Trans Assoc Comput Linguist. 2014; 2:207–218.62. Kiros R, Salakhutdinov R, Zemel RS. Unifying visual-semantic embeddings with multimodal neural language models. ArXiv. org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1411.2539.63. Donahue J, Hendricks LA, Rohrbach M, Venugopalan S, Guadarrama S, Saenko K, et al. Long-term recurrent convolutional networks for visual recognition and description. IEEE Trans Pattern Anal Mach Intell. 2017; 39:677–691.64. In : Vinyals O, Toshev A, Bengio S, Erhan D, editors. Show and tell: a neural image caption generator. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 3156–3164.65. In : Fang H, Gupta S, Iandola F, Srivastava R, Deng L, Dollár P, editors. From captions to visual concepts and back. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 1473–1482.66. Chen X, Zitnick CL. Mind's eye: a recurrent visual representation for image caption generation. In : Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 2422–2431.67. Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994; 5:157–166.68. Karpathy A, Li FF. Deep visual-semantic alignments for generating image descriptions. IEEE Trans Pattern Anal Mach Intell. 2016; 39:664–676.69. Hodosh M, Young P, Hockenmaier J. Framing image description as a ranking task: data, models and evaluation metrics. J Artif Intell Res. 2013; 47:853–899.70. Young P, Lai A, Hodosh M, Hockenmaier J. From image descriptions to visual denotations: new similarity metrics for semantic inference over event descriptions. Trans Assoc Comput Linguist. 2014; 2:67–78.71. Lin TY, Maire M, Belongie S, Hays J, Perona P, Ramanan D, et al. Microsoft COCO: common objects in context. In : Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. Computer Vision-ECCV 2014. New York. Springer;2014. p. 740–755.72. Shi Y, Suk HI, Gao Y, Shen D. Joint coupled-feature representation and coupled boosting for AD diagnosis. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2014; 2014:2721–2728.73. In : Hofmanninger J, Langs G, editors. Mapping visual features to semantic profiles for retrieval in medical imaging. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 457–465.74. In : Subbanna N, Precup D, Arbel T, editors. Iterative multilevel MRF leveraging context and voxel information for brain tumour segmentation in MRI. Proceedings of the 27th IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2014); 2014 June 23-28; Columbus, OH, USA. IEEE;2014. p. 400–405.75. In : Ngo TA, Carneiro G, editors. Fully automated non-rigid segmentation with distance regularized level set evolution initialized and constrained by deep-structured inference. Proceedings of 2014 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2014 June 23-28; Columbus, OH, USA. IEEE;2014. p. 3118–3125.76. In : Ledig C, Shi W, Bai W, Rueckert D, editors. Patch-based evaluation of image segmentation. Proceedings of 2014 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2014 June 23-28; Columbus, OH, USA. IEEE;2014. p. 3065–3072.77. In : Rupprecht C, Peter L, Navab N, editors. Image segmentation in twenty questions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 3314–3322.78. Kulkarni G, Premraj V, Ordonez V, Dhar S, Li S, Choi Y, et al. Babytalk: understanding and generating simple image descriptions. IEEE Trans Pattern Anal Mach Intell. 2013; 35:2891–2903.79. In : Feng Y, Lapata M, editors. How many words is a picture worth? Automatic caption generation for news images. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics; 2010 July 11-16; Uppsala, Sweden. Stroudsburg: Association for Computational Linguistics;2010. p. 1239–1249.80. Farhadi A, Hejrati M, Sadeghi MA, Young P, Rashtchian C, Hockenmaier J, et al. Every picture tells a story: generating sentences from images. In : Daniilidis K, Maragos P, Paragios N, editors. Computer Vision-ECCV 2010. Berlin, Heidelberg. Springer;2010. p. 15–29.81. In : Shin HC, Roberts K, Lu L, Fushman DD, Yao J, Summers RM, editors. Learning to read chest X-rays: recurrent neural cascade model for automated image annotation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016 June 27-30; Las Vegas, NV, USA. IEEE;2016.82. Bogoni L, Ko JP, Alpert J, Anand V, Fantauzzi J, Florin CH, et al. Impact of a computer-aided detection (CAD) system integrated into a picture archiving and communication system (PACS) on reader sensitivity and efficiency for the detection of lung nodules in thoracic CT exams. J Digit Imaging. 2012; 25:771–781.83. Welter P, Hocken C, Deserno TM, Grouls C, Günther RW. Workflow management of content-based image retrieval for CAD support in PACS environments based on IHE. Int J Comput Assist Radiol Surg. 2010; 5:393–400.84. Le AH, Liu B, Huang HK. Integration of computer-aided diagnosis/detection (CAD) results in a PACS environment using CAD-PACS toolkit and DICOM SR. Int J Comput Assist Radiol Surg. 2009; 4:317–329.85. Zhou Z. Data security assurance in CAD-PACS integration. Comput Med Imaging Graph. 2007; 31:353–360.86. Wang D, Khosla A, Gargeya R, Irshad H, Beck AH. Deep learning for identifying metastatic breast cancer. ArXiv.org Web site. Accessed April 1, 2017. https://arxiv.org/pdf/1606.05718.pdf.87. Hua KL, Hsu CH, Hidayati SC, Cheng WH, Chen YJ. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther. 2015; 8:2015–2022.88. In : Kumar D, Wong A, Clausi DA, editors. Lung Nodule Classification Using Deep Features in CT Images. Proceedings of 2015 12th Conference on Computer and Robot Vision; 2015 June 3-5; Halifax, Canada. IEEE;2015. p. 133–138.89. Suk HI, Lee SW, Shen D. Alzheimer's Disease Neuroimaging Initiative. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage. 2014; 101:569–582.90. Suk HI, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv. 2013; 16(Pt 2):583–590.91. In : Liu S, Lis S, Cai W, Pujol S, Kikinis R, Feng D, editors. Early diagnosis of Alzheimer's disease with deep learning. Proceedings of the IEEE 11th International Symposium on Biodmedical Imaging; 2014 April 29-May 2; Beijing, China. IEEE;2014. p. 1015–1018.92. Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep. 2016; 6:24454.93. Kallenberg M, Petersen K, Nielsen M, Ng AY, Pengfei Diao, Igel C, et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging. 2016; 35:1322–1331.94. Chen J, Chen J, Ding HY, Pan QS, Hong WD, Xu G, et al. Use of an artificial neural network to construct a model of predicting deep fungal infection in lung cancer patients. Asian Pac J Cancer Prev. 2015; 16:5095–5099.95. Capterra.com Web site. Spantel. SpeechRite. Accessed April 1, 2017. http://www.capterra.com/speech-recognition-software/spotlight/142035/SpeechRite/Spantel.96. 2ascribe.com Web site. Radiology Dictation & Transcription. Accessed April 1, 2017. https://www.2ascribe.com/transcription-services/radiology-dictation-transcription.97. Liu Y, Wang J. PACS and digital medicine: essential principles and modern practice. 1st ed. Boca Raton: CRC Press;2010.98. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015; 372:793–795.99. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014; 5:4006.100. Hesketh RL, Zhu AX, Oklu R. Radiomics and circulating tumor cells: personalized care in hepatocellular carcinoma? Diagn Interv Radiol. 2015; 21:78–84.101. Scikit-learn.org Web site. Scikit-learn algorithm cheat-sheet. Accessed April 1, 2017. http://scikit-learn.org/stable/tutorial/machine_learning_map/.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Deep Learning in Nuclear Medicine and Molecular Imaging: Current Perspectives and Future Directions
- Deep Learning in Dental Radiographic Imaging
- Current status of deep learning applications in abdominal ultrasonography
- Basics of Deep Learning: A Radiologist's Guide to Understanding Published Radiology Articles on Deep Learning
- Development of an Optimized Deep Learning Model for Medical Imaging